Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
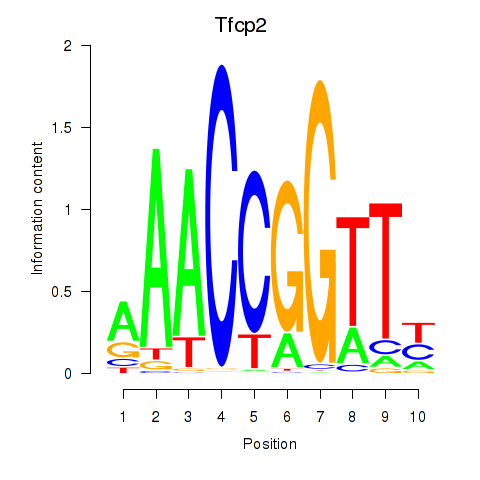
Results for Tfcp2
Z-value: 0.94
Transcription factors associated with Tfcp2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfcp2
|
ENSMUSG00000009733.10 | transcription factor CP2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfcp2 | mm39_v1_chr15_-_100449839_100449892 | 0.77 | 1.3e-01 | Click! |
Activity profile of Tfcp2 motif
Sorted Z-values of Tfcp2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_23206001 | 0.87 |
ENSMUST00000020538.13
ENSMUST00000109551.8 ENSMUST00000102870.8 ENSMUST00000102869.8 |
Xpo1
|
exportin 1 |
| chr17_-_34218301 | 0.86 |
ENSMUST00000235463.2
|
H2-K1
|
histocompatibility 2, K1, K region |
| chrX_+_138464065 | 0.65 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr11_-_48762170 | 0.53 |
ENSMUST00000049519.4
ENSMUST00000097271.4 |
Irgm1
|
immunity-related GTPase family M member 1 |
| chr12_+_24758968 | 0.51 |
ENSMUST00000154588.2
|
Rrm2
|
ribonucleotide reductase M2 |
| chr12_-_54703281 | 0.48 |
ENSMUST00000056228.8
|
Sptssa
|
serine palmitoyltransferase, small subunit A |
| chr10_+_43455919 | 0.47 |
ENSMUST00000214476.2
|
Cd24a
|
CD24a antigen |
| chr16_-_95928804 | 0.40 |
ENSMUST00000233292.2
ENSMUST00000050884.16 |
Hmgn1
|
high mobility group nucleosomal binding domain 1 |
| chr19_-_40576817 | 0.34 |
ENSMUST00000175932.2
ENSMUST00000176955.8 ENSMUST00000149476.3 |
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr19_-_40576782 | 0.34 |
ENSMUST00000176939.8
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr15_-_79976016 | 0.34 |
ENSMUST00000185306.3
|
Rpl3
|
ribosomal protein L3 |
| chr7_+_100970435 | 0.33 |
ENSMUST00000210192.2
ENSMUST00000172630.8 |
Stard10
|
START domain containing 10 |
| chr18_+_11766333 | 0.33 |
ENSMUST00000115861.9
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chr10_+_43455157 | 0.32 |
ENSMUST00000058714.10
|
Cd24a
|
CD24a antigen |
| chr8_+_70275079 | 0.32 |
ENSMUST00000164890.8
ENSMUST00000034325.6 ENSMUST00000238452.2 |
Lpar2
|
lysophosphatidic acid receptor 2 |
| chr8_+_86219191 | 0.32 |
ENSMUST00000034136.12
|
Gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr3_-_90603013 | 0.31 |
ENSMUST00000069960.12
ENSMUST00000117167.2 |
S100a9
|
S100 calcium binding protein A9 (calgranulin B) |
| chr8_-_61407760 | 0.31 |
ENSMUST00000110302.8
|
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr19_-_40576897 | 0.31 |
ENSMUST00000025979.13
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr4_+_89055359 | 0.31 |
ENSMUST00000058030.10
|
Mtap
|
methylthioadenosine phosphorylase |
| chr1_+_151220222 | 0.30 |
ENSMUST00000023918.13
ENSMUST00000111887.10 ENSMUST00000097543.8 |
Ivns1abp
|
influenza virus NS1A binding protein |
| chr11_+_68936457 | 0.30 |
ENSMUST00000108666.8
ENSMUST00000021277.6 |
Aurkb
|
aurora kinase B |
| chr17_+_35278011 | 0.30 |
ENSMUST00000007255.13
ENSMUST00000174493.8 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr6_+_134617903 | 0.29 |
ENSMUST00000062755.10
|
Borcs5
|
BLOC-1 related complex subunit 5 |
| chr17_+_35191661 | 0.28 |
ENSMUST00000007248.5
|
Hspa1l
|
heat shock protein 1-like |
| chr1_-_60137263 | 0.28 |
ENSMUST00000143342.8
|
Wdr12
|
WD repeat domain 12 |
| chr9_-_107215504 | 0.27 |
ENSMUST00000118051.2
ENSMUST00000035196.14 |
Hemk1
|
HemK methyltransferase family member 1 |
| chr13_-_100922910 | 0.27 |
ENSMUST00000174038.2
ENSMUST00000091295.14 ENSMUST00000072119.15 |
Ccnb1
|
cyclin B1 |
| chr19_-_6886898 | 0.26 |
ENSMUST00000238095.2
|
Prdx5
|
peroxiredoxin 5 |
| chr19_-_24178000 | 0.26 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2 |
| chr12_-_79211820 | 0.26 |
ENSMUST00000162569.8
|
Vti1b
|
vesicle transport through interaction with t-SNAREs 1B |
| chr11_+_103857541 | 0.26 |
ENSMUST00000057921.10
ENSMUST00000063347.12 |
Arf2
|
ADP-ribosylation factor 2 |
| chr6_-_34294377 | 0.26 |
ENSMUST00000154655.2
ENSMUST00000102980.11 |
Akr1b3
|
aldo-keto reductase family 1, member B3 (aldose reductase) |
| chr1_-_149836974 | 0.25 |
ENSMUST00000190507.2
ENSMUST00000070200.15 |
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr11_-_102588536 | 0.25 |
ENSMUST00000164506.3
ENSMUST00000092569.13 |
Ccdc43
|
coiled-coil domain containing 43 |
| chr1_-_52230062 | 0.24 |
ENSMUST00000156887.8
ENSMUST00000129107.2 |
Gls
|
glutaminase |
| chr19_-_8690330 | 0.24 |
ENSMUST00000206598.2
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr5_-_9047324 | 0.23 |
ENSMUST00000003720.5
|
Crot
|
carnitine O-octanoyltransferase |
| chr19_-_38032006 | 0.23 |
ENSMUST00000172095.3
ENSMUST00000041475.16 |
Myof
|
myoferlin |
| chr11_+_52122836 | 0.23 |
ENSMUST00000037324.12
ENSMUST00000166537.8 |
Skp1
|
S-phase kinase-associated protein 1 |
| chr19_-_46917661 | 0.23 |
ENSMUST00000236727.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr16_+_20430335 | 0.22 |
ENSMUST00000115522.10
ENSMUST00000119224.8 ENSMUST00000120394.8 ENSMUST00000079600.12 |
Eef1akmt4
Gm49333
|
EEF1A lysine methyltransferase 4 predicted gene, 49333 |
| chr5_-_34794185 | 0.22 |
ENSMUST00000149657.5
|
Mfsd10
|
major facilitator superfamily domain containing 10 |
| chr11_+_3845221 | 0.22 |
ENSMUST00000109996.8
ENSMUST00000055931.5 |
Dusp18
|
dual specificity phosphatase 18 |
| chr11_+_50267808 | 0.22 |
ENSMUST00000109142.8
|
Hnrnph1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr11_-_70860778 | 0.22 |
ENSMUST00000108530.2
ENSMUST00000035283.11 ENSMUST00000108531.8 |
Nup88
|
nucleoporin 88 |
| chr8_+_34089597 | 0.22 |
ENSMUST00000009774.11
|
Ppp2cb
|
protein phosphatase 2 (formerly 2A), catalytic subunit, beta isoform |
| chr14_-_59632830 | 0.21 |
ENSMUST00000166912.3
|
Phf11c
|
PHD finger protein 11C |
| chr19_-_38031774 | 0.21 |
ENSMUST00000226068.2
|
Myof
|
myoferlin |
| chr7_+_100970910 | 0.21 |
ENSMUST00000174291.8
ENSMUST00000167888.9 ENSMUST00000172662.2 |
Stard10
|
START domain containing 10 |
| chr9_+_50768224 | 0.21 |
ENSMUST00000174628.8
ENSMUST00000034560.14 ENSMUST00000114437.9 ENSMUST00000175645.8 ENSMUST00000176349.8 ENSMUST00000176798.8 ENSMUST00000175640.8 |
Ppp2r1b
|
protein phosphatase 2, regulatory subunit A, beta |
| chr1_-_161078723 | 0.20 |
ENSMUST00000051925.5
ENSMUST00000071718.12 |
Prdx6
|
peroxiredoxin 6 |
| chr5_+_129864044 | 0.20 |
ENSMUST00000201414.5
|
Cct6a
|
chaperonin containing Tcp1, subunit 6a (zeta) |
| chr11_-_51541610 | 0.19 |
ENSMUST00000142721.2
ENSMUST00000156835.8 ENSMUST00000001080.16 |
N4bp3
|
NEDD4 binding protein 3 |
| chr1_-_30988772 | 0.19 |
ENSMUST00000238874.2
ENSMUST00000027232.15 ENSMUST00000076587.6 ENSMUST00000233506.2 |
Ptp4a1
|
protein tyrosine phosphatase 4a1 |
| chr8_-_27618643 | 0.19 |
ENSMUST00000033877.6
|
Brf2
|
BRF2, RNA polymerase III transcription initiation factor 50kDa subunit |
| chr14_-_70945434 | 0.19 |
ENSMUST00000228346.2
|
Xpo7
|
exportin 7 |
| chr7_-_84661476 | 0.19 |
ENSMUST00000124773.3
ENSMUST00000233725.2 ENSMUST00000233739.2 ENSMUST00000232837.2 |
Vmn2r66
|
vomeronasal 2, receptor 66 |
| chr1_+_16735401 | 0.19 |
ENSMUST00000177501.2
ENSMUST00000065373.6 |
Tmem70
|
transmembrane protein 70 |
| chr16_+_20492267 | 0.18 |
ENSMUST00000115460.8
|
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr17_+_25992761 | 0.18 |
ENSMUST00000237541.2
|
Ciao3
|
cytosolic iron-sulfur assembly component 3 |
| chr19_+_8907206 | 0.18 |
ENSMUST00000224272.2
|
Eml3
|
echinoderm microtubule associated protein like 3 |
| chrX_-_162859095 | 0.18 |
ENSMUST00000037928.9
|
Siah1b
|
siah E3 ubiquitin protein ligase 1B |
| chrX_-_162859126 | 0.18 |
ENSMUST00000071667.9
|
Siah1b
|
siah E3 ubiquitin protein ligase 1B |
| chr9_+_4376556 | 0.18 |
ENSMUST00000212075.2
|
Msantd4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chr5_+_107112186 | 0.17 |
ENSMUST00000117196.9
ENSMUST00000031221.12 ENSMUST00000076467.13 |
Cdc7
|
cell division cycle 7 (S. cerevisiae) |
| chr4_-_139079609 | 0.17 |
ENSMUST00000030513.13
ENSMUST00000155257.8 |
Mrto4
|
mRNA turnover 4, ribosome maturation factor |
| chr2_-_5017526 | 0.17 |
ENSMUST00000027980.8
|
Mcm10
|
minichromosome maintenance 10 replication initiation factor |
| chr3_-_95014090 | 0.17 |
ENSMUST00000005768.8
ENSMUST00000107232.9 ENSMUST00000107236.9 |
Pip5k1a
|
phosphatidylinositol-4-phosphate 5-kinase, type 1 alpha |
| chr8_+_94899292 | 0.17 |
ENSMUST00000034214.8
ENSMUST00000212806.2 |
Mt2
|
metallothionein 2 |
| chr14_-_54651442 | 0.17 |
ENSMUST00000227334.2
|
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr11_-_102207486 | 0.16 |
ENSMUST00000146896.9
ENSMUST00000079589.11 |
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr4_-_150087587 | 0.16 |
ENSMUST00000084117.13
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr13_-_25121568 | 0.16 |
ENSMUST00000037615.7
|
Aldh5a1
|
aldhehyde dehydrogenase family 5, subfamily A1 |
| chrX_+_36059274 | 0.16 |
ENSMUST00000016463.4
|
Slc25a5
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 5 |
| chr6_-_47790272 | 0.16 |
ENSMUST00000077290.9
|
Pdia4
|
protein disulfide isomerase associated 4 |
| chr1_-_43866910 | 0.16 |
ENSMUST00000153317.6
ENSMUST00000128261.2 ENSMUST00000126008.8 ENSMUST00000139451.8 |
Uxs1
|
UDP-glucuronate decarboxylase 1 |
| chr11_-_80032949 | 0.16 |
ENSMUST00000131601.2
ENSMUST00000050207.10 |
Tefm
|
transcription elongation factor, mitochondrial |
| chr8_-_111573401 | 0.16 |
ENSMUST00000042012.7
|
Sf3b3
|
splicing factor 3b, subunit 3 |
| chr5_+_33815466 | 0.15 |
ENSMUST00000074849.13
ENSMUST00000079534.11 ENSMUST00000201633.2 |
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr7_+_29794575 | 0.15 |
ENSMUST00000130526.2
ENSMUST00000108200.2 |
Zfp260
|
zinc finger protein 260 |
| chr4_-_149858694 | 0.15 |
ENSMUST00000105686.3
|
Slc25a33
|
solute carrier family 25, member 33 |
| chr17_-_26051871 | 0.15 |
ENSMUST00000236515.2
ENSMUST00000236361.2 |
Stub1
|
STIP1 homology and U-Box containing protein 1 |
| chr6_+_116185077 | 0.15 |
ENSMUST00000204051.2
|
Washc2
|
WASH complex subunit 2` |
| chr11_-_17161504 | 0.14 |
ENSMUST00000020317.8
|
Pno1
|
partner of NOB1 homolog |
| chr2_-_174280811 | 0.14 |
ENSMUST00000016400.9
|
Ctsz
|
cathepsin Z |
| chr11_+_48691175 | 0.14 |
ENSMUST00000020640.8
|
Rack1
|
receptor for activated C kinase 1 |
| chr1_-_85087567 | 0.14 |
ENSMUST00000161675.3
ENSMUST00000160792.8 |
A530032D15Rik
|
RIKEN cDNA A530032D15Rik gene |
| chr6_+_67838100 | 0.14 |
ENSMUST00000200586.2
ENSMUST00000103309.3 |
Igkv17-127
|
immunoglobulin kappa variable 17-127 |
| chrX_+_163763588 | 0.14 |
ENSMUST00000167446.8
ENSMUST00000057150.8 |
Fancb
|
Fanconi anemia, complementation group B |
| chr3_-_95014218 | 0.14 |
ENSMUST00000107233.9
|
Pip5k1a
|
phosphatidylinositol-4-phosphate 5-kinase, type 1 alpha |
| chr4_-_133695204 | 0.14 |
ENSMUST00000100472.10
ENSMUST00000136327.2 |
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr11_+_54757239 | 0.14 |
ENSMUST00000117710.2
|
Hint1
|
histidine triad nucleotide binding protein 1 |
| chr19_-_60849828 | 0.14 |
ENSMUST00000135808.8
|
Sfxn4
|
sideroflexin 4 |
| chr5_+_114924106 | 0.14 |
ENSMUST00000137519.2
|
Ankrd13a
|
ankyrin repeat domain 13a |
| chr15_+_79113341 | 0.13 |
ENSMUST00000163571.8
|
Pick1
|
protein interacting with C kinase 1 |
| chr1_-_133617824 | 0.13 |
ENSMUST00000189524.2
ENSMUST00000169295.8 |
Lax1
|
lymphocyte transmembrane adaptor 1 |
| chrX_-_74174524 | 0.13 |
ENSMUST00000114091.8
|
Mpp1
|
membrane protein, palmitoylated |
| chr11_-_82719850 | 0.13 |
ENSMUST00000021036.13
ENSMUST00000074515.11 ENSMUST00000103218.3 |
Rffl
|
ring finger and FYVE like domain containing protein |
| chr7_-_44983080 | 0.13 |
ENSMUST00000211743.2
ENSMUST00000042194.10 |
Trpm4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr8_+_57964956 | 0.13 |
ENSMUST00000210871.2
|
Hmgb2
|
high mobility group box 2 |
| chr2_-_30249202 | 0.13 |
ENSMUST00000100215.11
ENSMUST00000113620.10 ENSMUST00000163668.3 ENSMUST00000028214.15 ENSMUST00000113621.10 |
Sh3glb2
|
SH3-domain GRB2-like endophilin B2 |
| chr10_+_13376745 | 0.13 |
ENSMUST00000060212.13
ENSMUST00000121465.3 |
Fuca2
|
fucosidase, alpha-L- 2, plasma |
| chr15_-_96597610 | 0.13 |
ENSMUST00000023099.8
|
Slc38a2
|
solute carrier family 38, member 2 |
| chr9_+_108368032 | 0.13 |
ENSMUST00000166103.9
ENSMUST00000085044.14 ENSMUST00000193678.6 ENSMUST00000178075.8 |
Usp19
|
ubiquitin specific peptidase 19 |
| chr1_-_85087170 | 0.13 |
ENSMUST00000161724.8
|
A530032D15Rik
|
RIKEN cDNA A530032D15Rik gene |
| chr4_+_109533753 | 0.13 |
ENSMUST00000102724.5
|
Faf1
|
Fas-associated factor 1 |
| chr5_+_124767114 | 0.13 |
ENSMUST00000037865.13
|
Atp6v0a2
|
ATPase, H+ transporting, lysosomal V0 subunit A2 |
| chr3_-_143908060 | 0.12 |
ENSMUST00000121112.6
|
Lmo4
|
LIM domain only 4 |
| chr10_+_34173426 | 0.12 |
ENSMUST00000047935.8
|
Tspyl4
|
TSPY-like 4 |
| chrX_+_52001108 | 0.12 |
ENSMUST00000078944.13
ENSMUST00000101587.10 ENSMUST00000154864.4 |
Phf6
|
PHD finger protein 6 |
| chr4_-_133329479 | 0.12 |
ENSMUST00000057311.4
|
Sfn
|
stratifin |
| chr15_-_75793312 | 0.12 |
ENSMUST00000053918.9
|
Pycrl
|
pyrroline-5-carboxylate reductase-like |
| chr10_+_81104006 | 0.12 |
ENSMUST00000057798.9
ENSMUST00000219304.2 |
Apba3
|
amyloid beta (A4) precursor protein-binding, family A, member 3 |
| chr1_+_55091922 | 0.11 |
ENSMUST00000087617.11
ENSMUST00000027125.7 |
Coq10b
|
coenzyme Q10B |
| chr2_+_84629172 | 0.11 |
ENSMUST00000102642.9
ENSMUST00000150325.2 |
Ube2l6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr19_-_45738002 | 0.11 |
ENSMUST00000070215.8
|
Npm3
|
nucleoplasmin 3 |
| chr4_-_139079842 | 0.11 |
ENSMUST00000102503.10
|
Mrto4
|
mRNA turnover 4, ribosome maturation factor |
| chr2_+_129642371 | 0.11 |
ENSMUST00000165413.9
ENSMUST00000166282.3 |
Stk35
|
serine/threonine kinase 35 |
| chr3_+_84859453 | 0.11 |
ENSMUST00000029727.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr13_-_100152069 | 0.11 |
ENSMUST00000022148.7
|
Mccc2
|
methylcrotonoyl-Coenzyme A carboxylase 2 (beta) |
| chr4_+_133311665 | 0.10 |
ENSMUST00000030661.14
ENSMUST00000105899.2 |
Gpn2
|
GPN-loop GTPase 2 |
| chr12_-_114252202 | 0.10 |
ENSMUST00000195124.6
ENSMUST00000103481.3 |
Ighv3-6
|
immunoglobulin heavy variable 3-6 |
| chr2_+_91033230 | 0.10 |
ENSMUST00000150403.8
ENSMUST00000002172.14 ENSMUST00000238832.2 ENSMUST00000239169.2 ENSMUST00000155418.2 |
Acp2
|
acid phosphatase 2, lysosomal |
| chr11_+_4852212 | 0.10 |
ENSMUST00000142543.3
|
Thoc5
|
THO complex 5 |
| chr11_-_61065846 | 0.10 |
ENSMUST00000041683.9
|
Usp22
|
ubiquitin specific peptidase 22 |
| chr11_-_5691117 | 0.10 |
ENSMUST00000140922.2
ENSMUST00000093362.12 |
Urgcp
|
upregulator of cell proliferation |
| chr3_-_143908111 | 0.10 |
ENSMUST00000121796.8
|
Lmo4
|
LIM domain only 4 |
| chr12_+_78908466 | 0.10 |
ENSMUST00000071230.8
|
Eif2s1
|
eukaryotic translation initiation factor 2, subunit 1 alpha |
| chr2_+_32460868 | 0.10 |
ENSMUST00000140592.8
ENSMUST00000028151.7 |
Dpm2
|
dolichol-phosphate (beta-D) mannosyltransferase 2 |
| chr2_+_112092271 | 0.10 |
ENSMUST00000028553.4
|
Nop10
|
NOP10 ribonucleoprotein |
| chr5_-_24782465 | 0.10 |
ENSMUST00000030795.10
|
Abcf2
|
ATP-binding cassette, sub-family F (GCN20), member 2 |
| chr5_+_33815910 | 0.10 |
ENSMUST00000114426.10
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr7_+_100971034 | 0.10 |
ENSMUST00000173270.8
|
Stard10
|
START domain containing 10 |
| chr17_+_28426831 | 0.10 |
ENSMUST00000233264.2
|
Def6
|
differentially expressed in FDCP 6 |
| chr11_-_102207516 | 0.10 |
ENSMUST00000107115.8
ENSMUST00000128016.2 |
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chrX_-_163763273 | 0.09 |
ENSMUST00000112247.9
ENSMUST00000004715.2 |
Mospd2
|
motile sperm domain containing 2 |
| chr13_+_51562675 | 0.09 |
ENSMUST00000087978.5
|
S1pr3
|
sphingosine-1-phosphate receptor 3 |
| chr4_+_118478357 | 0.09 |
ENSMUST00000147373.2
|
Ebna1bp2
|
EBNA1 binding protein 2 |
| chr17_+_34812361 | 0.09 |
ENSMUST00000174532.2
|
Pbx2
|
pre B cell leukemia homeobox 2 |
| chr4_-_133695264 | 0.09 |
ENSMUST00000102553.11
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr7_+_136496328 | 0.09 |
ENSMUST00000081510.4
|
Mgmt
|
O-6-methylguanine-DNA methyltransferase |
| chr19_-_53932581 | 0.09 |
ENSMUST00000236885.2
ENSMUST00000236098.2 ENSMUST00000236370.2 |
Bbip1
|
BBSome interacting protein 1 |
| chr5_+_124621227 | 0.08 |
ENSMUST00000136567.2
|
Snrnp35
|
small nuclear ribonucleoprotein 35 (U11/U12) |
| chr12_-_65120674 | 0.08 |
ENSMUST00000220983.2
ENSMUST00000220730.2 ENSMUST00000021332.10 |
Fkbp3
|
FK506 binding protein 3 |
| chr1_-_194813843 | 0.08 |
ENSMUST00000075451.12
ENSMUST00000191775.2 |
Cr1l
|
complement component (3b/4b) receptor 1-like |
| chr15_+_79113563 | 0.08 |
ENSMUST00000018295.14
ENSMUST00000053926.12 |
Pick1
Gm49486
|
protein interacting with C kinase 1 predicted gene, 49486 |
| chr13_+_34148691 | 0.08 |
ENSMUST00000021843.13
|
Nqo2
|
N-ribosyldihydronicotinamide quinone reductase 2 |
| chr16_+_43960183 | 0.08 |
ENSMUST00000159514.8
ENSMUST00000161326.8 ENSMUST00000063520.15 ENSMUST00000063542.8 |
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr16_-_3725515 | 0.08 |
ENSMUST00000177221.2
ENSMUST00000177323.8 |
1700037C18Rik
|
RIKEN cDNA 1700037C18 gene |
| chr17_+_56312672 | 0.07 |
ENSMUST00000133998.8
|
Mpnd
|
MPN domain containing |
| chr19_-_34143437 | 0.07 |
ENSMUST00000025686.9
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr17_-_56312555 | 0.07 |
ENSMUST00000043785.8
|
Stap2
|
signal transducing adaptor family member 2 |
| chr1_+_37469220 | 0.07 |
ENSMUST00000114925.10
ENSMUST00000027285.13 ENSMUST00000144617.8 ENSMUST00000193979.6 ENSMUST00000118059.3 ENSMUST00000193713.2 |
Unc50
|
unc-50 homolog |
| chr6_-_43643093 | 0.07 |
ENSMUST00000114644.8
ENSMUST00000067888.14 |
Tpk1
|
thiamine pyrophosphokinase |
| chr12_+_103524690 | 0.07 |
ENSMUST00000187155.7
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr16_-_43959559 | 0.07 |
ENSMUST00000063661.13
ENSMUST00000114666.9 |
Atp6v1a
|
ATPase, H+ transporting, lysosomal V1 subunit A |
| chr19_+_8906916 | 0.06 |
ENSMUST00000096241.6
|
Eml3
|
echinoderm microtubule associated protein like 3 |
| chr1_+_58432021 | 0.06 |
ENSMUST00000050552.15
|
Bzw1
|
basic leucine zipper and W2 domains 1 |
| chr10_+_79795977 | 0.06 |
ENSMUST00000045247.9
|
Wdr18
|
WD repeat domain 18 |
| chr11_-_95805710 | 0.06 |
ENSMUST00000038343.7
|
B4galnt2
|
beta-1,4-N-acetyl-galactosaminyl transferase 2 |
| chr3_-_68911886 | 0.06 |
ENSMUST00000169064.8
|
Ift80
|
intraflagellar transport 80 |
| chr5_-_30278552 | 0.06 |
ENSMUST00000198095.2
ENSMUST00000196872.2 ENSMUST00000026846.11 |
Tyms
|
thymidylate synthase |
| chr9_+_108367801 | 0.06 |
ENSMUST00000006854.13
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr18_+_61688329 | 0.06 |
ENSMUST00000165123.8
|
Csnk1a1
|
casein kinase 1, alpha 1 |
| chr5_-_134717443 | 0.06 |
ENSMUST00000015137.10
|
Limk1
|
LIM-domain containing, protein kinase |
| chr1_+_165130192 | 0.06 |
ENSMUST00000111450.3
|
Gpr161
|
G protein-coupled receptor 161 |
| chr7_+_141047416 | 0.05 |
ENSMUST00000209988.2
|
Cd151
|
CD151 antigen |
| chr4_+_95467701 | 0.05 |
ENSMUST00000150830.2
ENSMUST00000134012.9 |
Fggy
|
FGGY carbohydrate kinase domain containing |
| chr1_+_34890912 | 0.05 |
ENSMUST00000152654.3
|
Plekhb2
|
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr2_-_84605732 | 0.05 |
ENSMUST00000023994.10
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chrX_+_151016224 | 0.05 |
ENSMUST00000112588.9
ENSMUST00000082177.13 |
Kdm5c
|
lysine (K)-specific demethylase 5C |
| chr10_-_89568106 | 0.05 |
ENSMUST00000020109.5
|
Actr6
|
ARP6 actin-related protein 6 |
| chr10_+_81104041 | 0.05 |
ENSMUST00000218742.2
|
Apba3
|
amyloid beta (A4) precursor protein-binding, family A, member 3 |
| chr16_-_50252703 | 0.05 |
ENSMUST00000066037.13
ENSMUST00000089399.11 ENSMUST00000089404.10 ENSMUST00000138166.8 |
Bbx
|
bobby sox HMG box containing |
| chr12_-_70160491 | 0.05 |
ENSMUST00000222237.2
|
Nin
|
ninein |
| chr2_+_37342248 | 0.05 |
ENSMUST00000148470.8
ENSMUST00000066055.10 ENSMUST00000112920.2 |
Rabgap1
|
RAB GTPase activating protein 1 |
| chr17_+_25992742 | 0.05 |
ENSMUST00000134108.8
ENSMUST00000002350.11 |
Ciao3
|
cytosolic iron-sulfur assembly component 3 |
| chr10_-_18421628 | 0.05 |
ENSMUST00000213153.2
|
Hebp2
|
heme binding protein 2 |
| chr4_+_95445731 | 0.04 |
ENSMUST00000079223.11
ENSMUST00000177394.8 |
Fggy
|
FGGY carbohydrate kinase domain containing |
| chr19_-_6886965 | 0.04 |
ENSMUST00000173091.2
|
Prdx5
|
peroxiredoxin 5 |
| chr17_-_71153283 | 0.04 |
ENSMUST00000156484.2
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr11_+_116423266 | 0.04 |
ENSMUST00000106386.8
ENSMUST00000145737.8 ENSMUST00000155102.8 ENSMUST00000063446.13 |
Sphk1
|
sphingosine kinase 1 |
| chr14_+_79718604 | 0.04 |
ENSMUST00000040131.13
|
Elf1
|
E74-like factor 1 |
| chr19_-_12742811 | 0.04 |
ENSMUST00000112933.2
|
Cntf
|
ciliary neurotrophic factor |
| chr17_+_28426752 | 0.04 |
ENSMUST00000002327.6
ENSMUST00000233560.2 ENSMUST00000233958.2 ENSMUST00000233170.2 |
Def6
|
differentially expressed in FDCP 6 |
| chr14_+_59716265 | 0.04 |
ENSMUST00000224893.2
|
Cab39l
|
calcium binding protein 39-like |
| chr3_+_41697046 | 0.04 |
ENSMUST00000120167.8
ENSMUST00000108065.9 ENSMUST00000146165.8 ENSMUST00000192193.6 ENSMUST00000119572.8 ENSMUST00000026867.14 ENSMUST00000026868.13 |
D3Ertd751e
|
DNA segment, Chr 3, ERATO Doi 751, expressed |
| chr8_+_105991280 | 0.04 |
ENSMUST00000036221.12
|
Fbxl8
|
F-box and leucine-rich repeat protein 8 |
| chr19_-_6887361 | 0.04 |
ENSMUST00000025904.12
|
Prdx5
|
peroxiredoxin 5 |
| chr11_-_100830183 | 0.04 |
ENSMUST00000092671.12
ENSMUST00000103114.8 |
Stat3
|
signal transducer and activator of transcription 3 |
| chr9_+_35334878 | 0.04 |
ENSMUST00000154652.8
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr2_-_84605764 | 0.03 |
ENSMUST00000111641.2
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr17_-_24746911 | 0.03 |
ENSMUST00000176652.8
|
Traf7
|
TNF receptor-associated factor 7 |
| chr10_+_69048464 | 0.03 |
ENSMUST00000020101.12
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr7_+_130633776 | 0.03 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr2_+_3705824 | 0.03 |
ENSMUST00000115054.9
|
Fam107b
|
family with sequence similarity 107, member B |
| chr14_-_21898992 | 0.03 |
ENSMUST00000124549.9
|
Comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr18_+_69654231 | 0.03 |
ENSMUST00000202350.4
ENSMUST00000202477.4 |
Tcf4
|
transcription factor 4 |
| chr18_+_37869950 | 0.03 |
ENSMUST00000091935.7
|
Pcdhga9
|
protocadherin gamma subfamily A, 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfcp2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 0.3 | 0.8 | GO:0034118 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 0.2 | 0.9 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.1 | 0.4 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.1 | 1.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.3 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.3 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 0.3 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 0.3 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.1 | 0.3 | GO:0090420 | hexitol metabolic process(GO:0006059) naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 0.1 | 0.3 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.1 | 0.5 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.2 | GO:0015904 | tetracycline transport(GO:0015904) |
| 0.1 | 0.3 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.1 | 0.6 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.1 | 0.2 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.2 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.0 | 0.1 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 0.0 | 0.2 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.3 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.3 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.3 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.1 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.2 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.6 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.1 | GO:0090035 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.0 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.3 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.3 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.2 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.2 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.1 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.1 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.2 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.7 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.1 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.2 | GO:1904867 | protein localization to nuclear body(GO:1903405) positive regulation of establishment of protein localization to telomere(GO:1904851) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.0 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.0 | 0.2 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.1 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.0 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 0.2 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.1 | 0.9 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.3 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.1 | 0.9 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.5 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.3 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.3 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.5 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.5 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.3 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0019202 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 0.1 | 0.3 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.5 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.5 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.3 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.1 | 1.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.2 | GO:0008493 | tetracycline transporter activity(GO:0008493) |
| 0.1 | 0.3 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.1 | 0.9 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.2 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.1 | 0.2 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.3 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.1 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.2 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.3 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.2 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.3 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.8 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:1904408 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.3 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |