Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
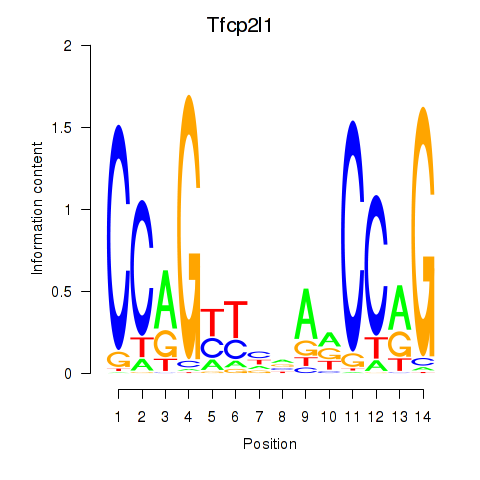
Results for Tfcp2l1
Z-value: 0.66
Transcription factors associated with Tfcp2l1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfcp2l1
|
ENSMUSG00000026380.11 | transcription factor CP2-like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfcp2l1 | mm39_v1_chr1_+_118555668_118555686 | 0.62 | 2.7e-01 | Click! |
Activity profile of Tfcp2l1 motif
Sorted Z-values of Tfcp2l1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_23947641 | 1.56 |
ENSMUST00000055770.4
|
H1f1
|
H1.1 linker histone, cluster member |
| chr19_+_8975249 | 0.75 |
ENSMUST00000236390.2
|
Ahnak
|
AHNAK nucleoprotein (desmoyokin) |
| chr14_+_25459690 | 0.47 |
ENSMUST00000007961.15
|
Zmiz1
|
zinc finger, MIZ-type containing 1 |
| chr16_-_76169902 | 0.47 |
ENSMUST00000054178.8
|
Nrip1
|
nuclear receptor interacting protein 1 |
| chr2_+_180829040 | 0.39 |
ENSMUST00000016488.13
ENSMUST00000108841.2 |
Ppdpf
|
pancreatic progenitor cell differentiation and proliferation factor |
| chr6_-_28831746 | 0.38 |
ENSMUST00000062304.7
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr19_-_24454720 | 0.36 |
ENSMUST00000099556.2
|
Fam122a
|
family with sequence similarity 122, member A |
| chr5_-_138262178 | 0.36 |
ENSMUST00000048421.14
|
Map11
|
microtubule associated protein 11 |
| chr14_+_25459630 | 0.33 |
ENSMUST00000162645.8
|
Zmiz1
|
zinc finger, MIZ-type containing 1 |
| chr15_+_102885467 | 0.33 |
ENSMUST00000001706.7
|
Hoxc9
|
homeobox C9 |
| chr16_-_76170714 | 0.32 |
ENSMUST00000231585.2
ENSMUST00000121927.8 |
Nrip1
|
nuclear receptor interacting protein 1 |
| chr9_+_106247943 | 0.30 |
ENSMUST00000173748.2
|
Dusp7
|
dual specificity phosphatase 7 |
| chr5_-_31065036 | 0.28 |
ENSMUST00000132034.5
ENSMUST00000132253.5 |
Ost4
|
oligosaccharyltransferase complex subunit 4 (non-catalytic) |
| chr8_-_72763462 | 0.28 |
ENSMUST00000003574.5
|
Cyp4f18
|
cytochrome P450, family 4, subfamily f, polypeptide 18 |
| chr2_+_90912710 | 0.26 |
ENSMUST00000169852.2
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr11_+_75422516 | 0.26 |
ENSMUST00000149727.8
ENSMUST00000108433.8 ENSMUST00000042561.14 ENSMUST00000143035.8 |
Slc43a2
|
solute carrier family 43, member 2 |
| chr11_+_77873535 | 0.22 |
ENSMUST00000108360.8
ENSMUST00000049167.14 |
Phf12
|
PHD finger protein 12 |
| chr2_-_26012751 | 0.22 |
ENSMUST00000140993.2
ENSMUST00000028300.6 |
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing |
| chr1_-_150341911 | 0.21 |
ENSMUST00000162367.8
ENSMUST00000161611.8 ENSMUST00000161320.8 ENSMUST00000159035.2 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr2_+_156562956 | 0.20 |
ENSMUST00000109566.9
ENSMUST00000146412.9 ENSMUST00000177013.8 ENSMUST00000171030.9 |
Dlgap4
|
DLG associated protein 4 |
| chr19_+_6468761 | 0.20 |
ENSMUST00000113462.8
ENSMUST00000077182.13 ENSMUST00000236635.2 ENSMUST00000113461.8 |
Nrxn2
|
neurexin II |
| chr11_+_68979308 | 0.19 |
ENSMUST00000021273.13
|
Vamp2
|
vesicle-associated membrane protein 2 |
| chr18_-_74197970 | 0.19 |
ENSMUST00000159162.2
ENSMUST00000091851.10 |
Mapk4
|
mitogen-activated protein kinase 4 |
| chr14_+_34542053 | 0.19 |
ENSMUST00000043349.7
|
Grid1
|
glutamate receptor, ionotropic, delta 1 |
| chr17_+_27171834 | 0.19 |
ENSMUST00000231853.2
|
Syngap1
|
synaptic Ras GTPase activating protein 1 homolog (rat) |
| chr6_-_90787100 | 0.19 |
ENSMUST00000101151.10
|
Iqsec1
|
IQ motif and Sec7 domain 1 |
| chr1_+_130659700 | 0.18 |
ENSMUST00000039323.8
|
AA986860
|
expressed sequence AA986860 |
| chr10_-_68114543 | 0.18 |
ENSMUST00000219238.2
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr10_-_80382611 | 0.16 |
ENSMUST00000183233.9
ENSMUST00000182604.8 |
Rexo1
|
REX1, RNA exonuclease 1 |
| chr13_+_81034214 | 0.16 |
ENSMUST00000161441.2
|
Arrdc3
|
arrestin domain containing 3 |
| chr13_-_115068626 | 0.16 |
ENSMUST00000056117.10
|
Itga2
|
integrin alpha 2 |
| chrX_+_101048650 | 0.16 |
ENSMUST00000144753.2
|
Nhsl2
|
NHS-like 2 |
| chr11_+_75422925 | 0.15 |
ENSMUST00000169547.9
|
Slc43a2
|
solute carrier family 43, member 2 |
| chr17_-_26417982 | 0.15 |
ENSMUST00000142410.2
ENSMUST00000120333.8 ENSMUST00000039113.14 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr3_-_107425316 | 0.15 |
ENSMUST00000169449.8
ENSMUST00000029499.15 |
Slc6a17
|
solute carrier family 6 (neurotransmitter transporter), member 17 |
| chr3_+_84500854 | 0.14 |
ENSMUST00000062623.4
|
Tigd4
|
tigger transposable element derived 4 |
| chr7_-_96981221 | 0.14 |
ENSMUST00000139582.9
|
Usp35
|
ubiquitin specific peptidase 35 |
| chr2_+_156562929 | 0.14 |
ENSMUST00000131157.9
|
Dlgap4
|
DLG associated protein 4 |
| chr5_+_30805265 | 0.13 |
ENSMUST00000062962.12
|
Slc35f6
|
solute carrier family 35, member F6 |
| chr2_+_127967951 | 0.13 |
ENSMUST00000089634.12
ENSMUST00000019281.14 ENSMUST00000110341.9 ENSMUST00000103211.8 ENSMUST00000103210.2 |
Bcl2l11
|
BCL2-like 11 (apoptosis facilitator) |
| chr15_+_80595486 | 0.13 |
ENSMUST00000067689.9
|
Tnrc6b
|
trinucleotide repeat containing 6b |
| chr4_+_118818775 | 0.13 |
ENSMUST00000058651.5
|
Lao1
|
L-amino acid oxidase 1 |
| chr2_+_156562989 | 0.12 |
ENSMUST00000000094.14
|
Dlgap4
|
DLG associated protein 4 |
| chr11_-_5331734 | 0.11 |
ENSMUST00000172492.8
|
Znrf3
|
zinc and ring finger 3 |
| chr6_+_97906760 | 0.10 |
ENSMUST00000101123.10
|
Mitf
|
melanogenesis associated transcription factor |
| chr14_-_55950939 | 0.10 |
ENSMUST00000168729.8
ENSMUST00000228123.2 ENSMUST00000178034.9 |
Tgm1
|
transglutaminase 1, K polypeptide |
| chrX_-_7185424 | 0.10 |
ENSMUST00000115746.8
|
Clcn5
|
chloride channel, voltage-sensitive 5 |
| chr7_+_96981517 | 0.08 |
ENSMUST00000054107.6
|
Kctd21
|
potassium channel tetramerisation domain containing 21 |
| chr7_-_100306160 | 0.08 |
ENSMUST00000107046.8
ENSMUST00000107045.9 ENSMUST00000139708.9 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr4_-_148021159 | 0.08 |
ENSMUST00000105712.2
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr4_-_135300934 | 0.08 |
ENSMUST00000105855.2
|
Grhl3
|
grainyhead like transcription factor 3 |
| chr4_-_148021217 | 0.08 |
ENSMUST00000019199.14
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr11_+_75422953 | 0.08 |
ENSMUST00000127226.3
|
Slc43a2
|
solute carrier family 43, member 2 |
| chr2_-_119916159 | 0.07 |
ENSMUST00000156159.4
|
Sptbn5
|
spectrin beta, non-erythrocytic 5 |
| chr19_+_26582450 | 0.07 |
ENSMUST00000176769.9
ENSMUST00000208163.2 ENSMUST00000025862.15 ENSMUST00000176030.8 |
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr5_-_143278619 | 0.06 |
ENSMUST00000212355.2
|
Zfp853
|
zinc finger protein 853 |
| chr12_+_102094977 | 0.06 |
ENSMUST00000159329.8
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chrX_-_7185529 | 0.06 |
ENSMUST00000128319.2
|
Clcn5
|
chloride channel, voltage-sensitive 5 |
| chr17_+_48080113 | 0.06 |
ENSMUST00000160373.8
ENSMUST00000159641.8 |
Tfeb
|
transcription factor EB |
| chr1_+_171157137 | 0.05 |
ENSMUST00000142063.8
ENSMUST00000129116.8 |
Dedd
|
death effector domain-containing |
| chr9_-_20553576 | 0.05 |
ENSMUST00000155301.8
|
Fbxl12
|
F-box and leucine-rich repeat protein 12 |
| chr6_+_129510145 | 0.05 |
ENSMUST00000204487.3
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr12_-_91351177 | 0.05 |
ENSMUST00000141429.8
|
Cep128
|
centrosomal protein 128 |
| chr13_+_109769294 | 0.05 |
ENSMUST00000135275.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr17_+_36134398 | 0.04 |
ENSMUST00000173493.8
ENSMUST00000173147.8 |
Flot1
|
flotillin 1 |
| chr6_-_83549399 | 0.04 |
ENSMUST00000206592.2
ENSMUST00000206400.2 |
Stambp
|
STAM binding protein |
| chr19_-_45619559 | 0.03 |
ENSMUST00000160718.9
|
Fbxw4
|
F-box and WD-40 domain protein 4 |
| chr1_-_132635042 | 0.03 |
ENSMUST00000043189.14
|
Nfasc
|
neurofascin |
| chr11_-_69768875 | 0.03 |
ENSMUST00000178597.3
|
Tmem95
|
transmembrane protein 95 |
| chr8_+_57964921 | 0.03 |
ENSMUST00000067925.8
|
Hmgb2
|
high mobility group box 2 |
| chr14_-_30973164 | 0.03 |
ENSMUST00000226565.2
ENSMUST00000022459.5 |
Phf7
|
PHD finger protein 7 |
| chr9_+_32283779 | 0.03 |
ENSMUST00000047334.10
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr4_+_116983973 | 0.03 |
ENSMUST00000134074.8
|
Dynlt4
|
dynein light chain Tctex-type 4 |
| chr2_+_164611812 | 0.03 |
ENSMUST00000088248.13
ENSMUST00000001439.7 |
Ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr1_+_180720666 | 0.02 |
ENSMUST00000085797.6
|
Lefty2
|
left-right determination factor 2 |
| chr2_+_79465761 | 0.02 |
ENSMUST00000111788.8
ENSMUST00000111784.9 |
Itprid2
|
ITPR interacting domain containing 2 |
| chr4_+_116983995 | 0.02 |
ENSMUST00000062206.3
|
Dynlt4
|
dynein light chain Tctex-type 4 |
| chr2_+_154393691 | 0.02 |
ENSMUST00000104928.2
|
Actl10
|
actin-like 10 |
| chr17_+_36134122 | 0.02 |
ENSMUST00000001569.15
ENSMUST00000174080.8 |
Flot1
|
flotillin 1 |
| chr7_+_144450260 | 0.02 |
ENSMUST00000033389.7
ENSMUST00000207229.2 |
Fgf15
|
fibroblast growth factor 15 |
| chr3_+_55149947 | 0.02 |
ENSMUST00000167204.8
ENSMUST00000054237.14 |
Dclk1
|
doublecortin-like kinase 1 |
| chr5_+_115568002 | 0.02 |
ENSMUST00000067168.9
|
Msi1
|
musashi RNA-binding protein 1 |
| chr9_-_57672124 | 0.02 |
ENSMUST00000215158.2
|
Clk3
|
CDC-like kinase 3 |
| chr2_+_121189091 | 0.02 |
ENSMUST00000000317.13
ENSMUST00000129130.3 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chr19_+_4189786 | 0.02 |
ENSMUST00000096338.5
|
Gpr152
|
G protein-coupled receptor 152 |
| chr9_+_65278979 | 0.02 |
ENSMUST00000239433.2
|
Ubap1l
|
ubiquitin-associated protein 1-like |
| chr15_-_97902576 | 0.02 |
ENSMUST00000023123.15
|
Col2a1
|
collagen, type II, alpha 1 |
| chr9_-_58065800 | 0.02 |
ENSMUST00000168864.4
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr10_+_76411474 | 0.02 |
ENSMUST00000001183.8
|
Ftcd
|
formiminotransferase cyclodeaminase |
| chr1_+_133109059 | 0.01 |
ENSMUST00000187285.7
|
Plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr6_+_113448388 | 0.01 |
ENSMUST00000058300.14
|
Il17rc
|
interleukin 17 receptor C |
| chr7_+_79392888 | 0.01 |
ENSMUST00000035622.8
|
Wdr93
|
WD repeat domain 93 |
| chr11_-_69712970 | 0.01 |
ENSMUST00000045771.7
|
Spem1
|
sperm maturation 1 |
| chr15_-_97902515 | 0.01 |
ENSMUST00000088355.12
|
Col2a1
|
collagen, type II, alpha 1 |
| chr11_-_3402355 | 0.01 |
ENSMUST00000077078.12
ENSMUST00000064364.3 |
Rnf185
|
ring finger protein 185 |
| chr4_+_106768662 | 0.01 |
ENSMUST00000030367.15
ENSMUST00000149926.8 |
Ssbp3
|
single-stranded DNA binding protein 3 |
| chr9_+_32305259 | 0.01 |
ENSMUST00000172015.3
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr13_-_99584091 | 0.01 |
ENSMUST00000223725.2
|
Map1b
|
microtubule-associated protein 1B |
| chr8_-_95405234 | 0.01 |
ENSMUST00000213043.2
|
Pllp
|
plasma membrane proteolipid |
| chr1_+_171156942 | 0.01 |
ENSMUST00000111299.8
ENSMUST00000064950.11 |
Dedd
|
death effector domain-containing |
| chr8_-_125161061 | 0.01 |
ENSMUST00000140012.8
|
Pgbd5
|
piggyBac transposable element derived 5 |
| chr19_+_16416664 | 0.01 |
ENSMUST00000237350.2
|
Gna14
|
guanine nucleotide binding protein, alpha 14 |
| chr18_+_74198173 | 0.01 |
ENSMUST00000066583.2
|
Gm9925
|
predicted gene 9925 |
| chr2_-_38604503 | 0.01 |
ENSMUST00000028084.5
|
Nr5a1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr1_-_132635078 | 0.01 |
ENSMUST00000187861.7
|
Nfasc
|
neurofascin |
| chr4_-_106585127 | 0.01 |
ENSMUST00000106770.8
ENSMUST00000145044.2 |
Mroh7
|
maestro heat-like repeat family member 7 |
| chr19_+_48194464 | 0.01 |
ENSMUST00000078880.6
|
Sorcs3
|
sortilin-related VPS10 domain containing receptor 3 |
| chr3_-_107424637 | 0.01 |
ENSMUST00000166892.2
|
Slc6a17
|
solute carrier family 6 (neurotransmitter transporter), member 17 |
| chr1_+_40720731 | 0.01 |
ENSMUST00000192345.2
|
Slc9a2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 2 |
| chr1_+_191873078 | 0.01 |
ENSMUST00000078470.12
ENSMUST00000110844.3 |
Kcnh1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
| chr7_+_105290203 | 0.01 |
ENSMUST00000210893.2
|
Gm45799
|
predicted gene 45799 |
| chr15_+_80118269 | 0.00 |
ENSMUST00000229138.2
ENSMUST00000166030.2 |
Mief1
|
mitochondrial elongation factor 1 |
| chr19_+_44748354 | 0.00 |
ENSMUST00000173346.4
|
Pax2
|
paired box 2 |
| chr11_-_69493567 | 0.00 |
ENSMUST00000138694.2
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr2_-_119060330 | 0.00 |
ENSMUST00000110820.3
|
Ppp1r14d
|
protein phosphatase 1, regulatory inhibitor subunit 14D |
| chr8_-_123087485 | 0.00 |
ENSMUST00000044123.2
|
Trhr2
|
thyrotropin releasing hormone receptor 2 |
| chr16_-_88571089 | 0.00 |
ENSMUST00000054223.4
|
2310057N15Rik
|
RIKEN cDNA 2310057N15 gene |
| chr9_+_118931532 | 0.00 |
ENSMUST00000165231.8
ENSMUST00000140326.8 |
Dlec1
|
deleted in lung and esophageal cancer 1 |
| chr8_-_68574179 | 0.00 |
ENSMUST00000178529.2
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr11_-_87780720 | 0.00 |
ENSMUST00000049743.2
|
Olfr462
|
olfactory receptor 462 |
| chr2_+_127183550 | 0.00 |
ENSMUST00000089673.10
ENSMUST00000179618.8 |
Astl
|
astacin-like metalloendopeptidase (M12 family) |
| chr3_+_103482591 | 0.00 |
ENSMUST00000090697.11
ENSMUST00000239027.2 |
Syt6
|
synaptotagmin VI |
| chr17_+_87224776 | 0.00 |
ENSMUST00000042172.7
|
Tmem247
|
transmembrane protein 247 |
| chr11_+_101221895 | 0.00 |
ENSMUST00000017316.7
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr18_+_36661198 | 0.00 |
ENSMUST00000237174.2
ENSMUST00000236124.2 ENSMUST00000236779.2 ENSMUST00000235181.2 ENSMUST00000074298.13 ENSMUST00000115694.3 ENSMUST00000236126.2 |
Slc4a9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chrX_+_51880056 | 0.00 |
ENSMUST00000101588.2
|
Ccdc160
|
coiled-coil domain containing 160 |
| chr8_+_13485451 | 0.00 |
ENSMUST00000167505.3
ENSMUST00000167071.9 |
Tmem255b
|
transmembrane protein 255B |
| chr10_+_77556417 | 0.00 |
ENSMUST00000092370.3
|
Krtap12-1
|
keratin associated protein 12-1 |
| chr7_-_81148019 | 0.00 |
ENSMUST00000207871.2
ENSMUST00000073406.12 ENSMUST00000144156.2 |
BC048679
|
cDNA sequence BC048679 |
| chr3_+_103482547 | 0.00 |
ENSMUST00000121834.8
|
Syt6
|
synaptotagmin VI |
| chr3_-_62512561 | 0.00 |
ENSMUST00000058535.6
|
Gpr149
|
G protein-coupled receptor 149 |
| chr2_+_127183838 | 0.00 |
ENSMUST00000156747.2
|
Astl
|
astacin-like metalloendopeptidase (M12 family) |
| chr11_+_49385362 | 0.00 |
ENSMUST00000071807.2
|
Olfr1385
|
olfactory receptor 1385 |
| chr1_-_93088614 | 0.00 |
ENSMUST00000043718.12
|
Mab21l4
|
mab-21-like 4 |
| chr9_-_37623573 | 0.00 |
ENSMUST00000104875.2
|
Olfr160
|
olfactory receptor 160 |
| chr3_+_94391676 | 0.00 |
ENSMUST00000198384.3
|
Celf3
|
CUGBP, Elav-like family member 3 |
| chr17_-_36013189 | 0.00 |
ENSMUST00000135078.2
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr11_-_120552001 | 0.00 |
ENSMUST00000150458.2
|
Notum
|
notum palmitoleoyl-protein carboxylesterase |
| chr4_-_88562696 | 0.00 |
ENSMUST00000105149.3
|
Ifna13
|
interferon alpha 13 |
| chr2_-_120119544 | 0.00 |
ENSMUST00000094665.5
|
Pla2g4d
|
phospholipase A2, group IVD |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfcp2l1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 0.8 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.3 | GO:0042374 | menaquinone metabolic process(GO:0009233) phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 0.2 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.0 | 0.2 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.2 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.2 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.2 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.1 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.2 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.4 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.2 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.2 | GO:0051971 | positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.4 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.1 | GO:1902477 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.8 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 0.0 | 0.1 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.2 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.0 | 0.8 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |