Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Thrb
Z-value: 0.35
Transcription factors associated with Thrb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Thrb
|
ENSMUSG00000021779.20 | thyroid hormone receptor beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Thrb | mm39_v1_chr14_-_4488167_4488189 | -0.28 | 6.5e-01 | Click! |
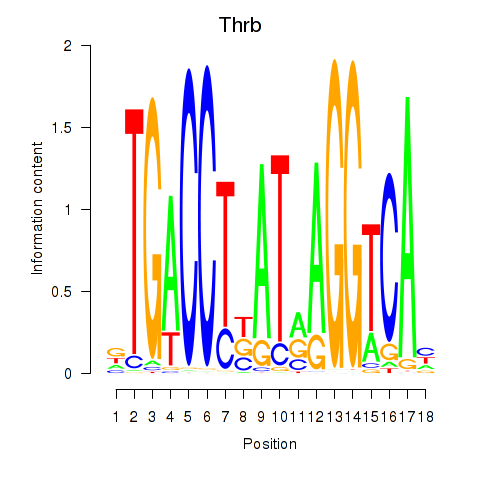
Activity profile of Thrb motif
Sorted Z-values of Thrb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_32253016 | 0.23 |
ENSMUST00000132028.8
ENSMUST00000136079.8 |
Ciz1
|
CDKN1A interacting zinc finger protein 1 |
| chr6_-_38814159 | 0.23 |
ENSMUST00000160962.8
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr5_-_30619246 | 0.20 |
ENSMUST00000114747.9
ENSMUST00000074171.10 |
Otof
|
otoferlin |
| chr15_-_103160082 | 0.17 |
ENSMUST00000149111.8
ENSMUST00000132836.8 |
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr9_+_102503476 | 0.15 |
ENSMUST00000190279.7
ENSMUST00000188398.7 |
Anapc13
|
anaphase promoting complex subunit 13 |
| chr11_+_46295547 | 0.10 |
ENSMUST00000063166.6
|
Fam71b
|
family with sequence similarity 71, member B |
| chrX_-_77627486 | 0.10 |
ENSMUST00000114025.8
ENSMUST00000134602.8 ENSMUST00000114024.9 |
Prrg1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr15_-_103159892 | 0.10 |
ENSMUST00000133600.8
ENSMUST00000134554.2 ENSMUST00000156927.8 |
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr10_-_18662526 | 0.09 |
ENSMUST00000216654.2
|
Gm4922
|
predicted gene 4922 |
| chr15_+_79230777 | 0.07 |
ENSMUST00000229130.2
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr11_+_49094292 | 0.07 |
ENSMUST00000150284.8
ENSMUST00000109197.8 ENSMUST00000151228.2 |
Zfp62
|
zinc finger protein 62 |
| chr7_-_24705320 | 0.07 |
ENSMUST00000102858.10
ENSMUST00000196684.2 ENSMUST00000080882.11 |
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr3_-_36529753 | 0.06 |
ENSMUST00000199478.2
|
Anxa5
|
annexin A5 |
| chr17_+_57297280 | 0.06 |
ENSMUST00000002735.9
|
Clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr4_-_148529187 | 0.05 |
ENSMUST00000051633.3
|
Ubiad1
|
UbiA prenyltransferase domain containing 1 |
| chrX_-_77627388 | 0.05 |
ENSMUST00000177904.8
|
Prrg1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr2_+_92213256 | 0.05 |
ENSMUST00000054316.9
ENSMUST00000111280.3 |
1700029I15Rik
|
RIKEN cDNA 1700029I15 gene |
| chr1_-_24626492 | 0.04 |
ENSMUST00000051344.6
ENSMUST00000115244.9 |
Col19a1
|
collagen, type XIX, alpha 1 |
| chr6_-_121450547 | 0.04 |
ENSMUST00000046373.8
ENSMUST00000151397.3 |
Iqsec3
|
IQ motif and Sec7 domain 3 |
| chr4_+_20042045 | 0.04 |
ENSMUST00000098242.4
|
Ggh
|
gamma-glutamyl hydrolase |
| chr1_-_83385911 | 0.04 |
ENSMUST00000160953.8
|
Sphkap
|
SPHK1 interactor, AKAP domain containing |
| chr9_+_102503815 | 0.04 |
ENSMUST00000038673.14
ENSMUST00000186693.2 |
Anapc13
|
anaphase promoting complex subunit 13 |
| chr11_+_100750316 | 0.04 |
ENSMUST00000107356.8
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr17_+_37253802 | 0.04 |
ENSMUST00000040498.12
|
Rnf39
|
ring finger protein 39 |
| chr8_-_120228434 | 0.03 |
ENSMUST00000133821.2
ENSMUST00000036748.15 |
Slc38a8
|
solute carrier family 38, member 8 |
| chr5_-_92426014 | 0.03 |
ENSMUST00000159345.8
ENSMUST00000113102.10 |
Naaa
|
N-acylethanolamine acid amidase |
| chr17_-_28908757 | 0.03 |
ENSMUST00000233398.2
|
Slc26a8
|
solute carrier family 26, member 8 |
| chr11_+_70350436 | 0.03 |
ENSMUST00000039093.10
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr12_+_108300599 | 0.03 |
ENSMUST00000021684.6
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr4_-_140573081 | 0.02 |
ENSMUST00000026378.4
|
Padi1
|
peptidyl arginine deiminase, type I |
| chr5_+_94365864 | 0.02 |
ENSMUST00000179743.3
|
Pramel38
|
PRAME like 38 |
| chr10_+_77522403 | 0.02 |
ENSMUST00000092366.4
|
Tspear
|
thrombospondin type laminin G domain and EAR repeats |
| chr3_+_101284391 | 0.02 |
ENSMUST00000043983.11
|
Igsf3
|
immunoglobulin superfamily, member 3 |
| chr14_+_53315909 | 0.01 |
ENSMUST00000103608.4
|
Trav14d-3-dv8
|
T cell receptor alpha variable 14D-3-DV8 |
| chr14_-_61597843 | 0.01 |
ENSMUST00000022494.10
|
Ebpl
|
emopamil binding protein-like |
| chr7_-_33065747 | 0.01 |
ENSMUST00000179248.2
|
Scgb2b20
|
secretoglobin, family 2B, member 20 |
| chr11_+_70350252 | 0.01 |
ENSMUST00000108563.9
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr11_-_58516867 | 0.01 |
ENSMUST00000203173.2
ENSMUST00000070804.3 |
Olfr323
|
olfactory receptor 323 |
| chr11_+_100750177 | 0.01 |
ENSMUST00000004145.14
ENSMUST00000133036.8 |
Stat5a
|
signal transducer and activator of transcription 5A |
| chr17_+_37253916 | 0.01 |
ENSMUST00000173072.2
|
Rnf39
|
ring finger protein 39 |
| chr9_+_44584523 | 0.01 |
ENSMUST00000034609.11
ENSMUST00000071219.12 |
Treh
|
trehalase (brush-border membrane glycoprotein) |
| chr7_+_27222678 | 0.01 |
ENSMUST00000108353.9
|
Hipk4
|
homeodomain interacting protein kinase 4 |
| chr18_+_77369654 | 0.01 |
ENSMUST00000096547.11
ENSMUST00000148341.9 ENSMUST00000123410.9 |
Loxhd1
|
lipoxygenase homology domains 1 |
| chr5_+_95021656 | 0.01 |
ENSMUST00000180076.2
|
Gm3183
|
predicted gene 3183 |
| chr14_+_54713703 | 0.01 |
ENSMUST00000164697.8
|
Rem2
|
rad and gem related GTP binding protein 2 |
| chr10_+_78786470 | 0.01 |
ENSMUST00000214952.2
ENSMUST00000203851.2 |
Olfr8
|
olfactory receptor 8 |
| chr14_+_54713557 | 0.01 |
ENSMUST00000164766.8
|
Rem2
|
rad and gem related GTP binding protein 2 |
| chr16_+_32238520 | 0.01 |
ENSMUST00000014220.15
ENSMUST00000080316.8 |
Dynlt2b
|
dynein light chain Tctex-type 2B |
| chr9_-_117843228 | 0.01 |
ENSMUST00000187803.3
|
Zcwpw2
|
zinc finger, CW type with PWWP domain 2 |
| chr7_-_31405182 | 0.00 |
ENSMUST00000178258.3
|
Scgb2b7
|
secretoglobin, family 2B, member 7 |
| chr10_+_87357657 | 0.00 |
ENSMUST00000020241.17
|
Pah
|
phenylalanine hydroxylase |
| chr2_-_36703102 | 0.00 |
ENSMUST00000100151.2
|
Olfr3
|
olfactory receptor 3 |
| chr7_-_32873289 | 0.00 |
ENSMUST00000186529.2
|
Scgb2b18
|
secretoglobin, family 2B, member 18 |
| chr7_-_31910570 | 0.00 |
ENSMUST00000188293.2
|
Scgb2b11
|
secretoglobin, family 2B, member 11 |
| chr13_-_59892731 | 0.00 |
ENSMUST00000180139.3
|
1700014D04Rik
|
RIKEN cDNA 1700014D04 gene |
| chr19_-_46314945 | 0.00 |
ENSMUST00000225781.2
ENSMUST00000223903.2 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr3_+_67281424 | 0.00 |
ENSMUST00000077916.12
|
Mlf1
|
myeloid leukemia factor 1 |
| chr5_+_35971697 | 0.00 |
ENSMUST00000130233.8
|
Ablim2
|
actin-binding LIM protein 2 |
| chr6_+_114435480 | 0.00 |
ENSMUST00000160780.2
|
Hrh1
|
histamine receptor H1 |
| chr9_-_107546195 | 0.00 |
ENSMUST00000192990.6
|
Slc38a3
|
solute carrier family 38, member 3 |
| chr2_+_58457370 | 0.00 |
ENSMUST00000071543.12
|
Upp2
|
uridine phosphorylase 2 |
| chr9_-_107546166 | 0.00 |
ENSMUST00000177567.8
|
Slc38a3
|
solute carrier family 38, member 3 |
| chr17_-_14223966 | 0.00 |
ENSMUST00000189454.3
|
Gm7356
|
predicted gene 7356 |
| chr8_+_105775224 | 0.00 |
ENSMUST00000093222.13
ENSMUST00000093223.5 |
Ces3a
|
carboxylesterase 3A |
| chr7_-_105249308 | 0.00 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr3_+_67281449 | 0.00 |
ENSMUST00000061322.10
|
Mlf1
|
myeloid leukemia factor 1 |
| chr6_+_70332836 | 0.00 |
ENSMUST00000103390.3
|
Igkv8-18
|
immunoglobulin kappa variable 8-18 |
| chr6_-_70217956 | 0.00 |
ENSMUST00000196275.2
|
Gm43218
|
predicted gene 43218 |
| chr2_-_25360043 | 0.00 |
ENSMUST00000114251.8
|
Ptgds
|
prostaglandin D2 synthase (brain) |
| chr7_-_32529087 | 0.00 |
ENSMUST00000182975.2
|
Scgb2b15
|
secretoglobin, family 2B, member 15 |
| chr5_+_130248547 | 0.00 |
ENSMUST00000202305.4
ENSMUST00000065329.13 ENSMUST00000200802.4 |
Tmem248
|
transmembrane protein 248 |
| chr2_-_25359752 | 0.00 |
ENSMUST00000114259.3
ENSMUST00000015234.13 |
Ptgds
|
prostaglandin D2 synthase (brain) |
| chr6_-_70364222 | 0.00 |
ENSMUST00000103392.3
ENSMUST00000195945.2 |
Igkv8-16
|
immunoglobulin kappa variable 8-16 |
| chr1_-_43203051 | 0.00 |
ENSMUST00000008280.14
|
Fhl2
|
four and a half LIM domains 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Thrb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.3 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 0.0 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.0 | GO:0060376 | positive regulation of mast cell differentiation(GO:0060376) |
| 0.0 | 0.1 | GO:0070459 | prolactin secretion(GO:0070459) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |