Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
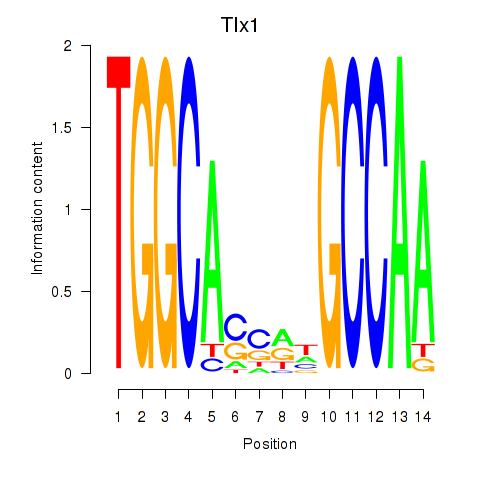
Results for Tlx1
Z-value: 0.58
Transcription factors associated with Tlx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tlx1
|
ENSMUSG00000025215.11 | T cell leukemia, homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tlx1 | mm39_v1_chr19_+_45139908_45140034 | 0.19 | 7.6e-01 | Click! |
Activity profile of Tlx1 motif
Sorted Z-values of Tlx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_42741148 | 0.64 |
ENSMUST00000236659.2
ENSMUST00000076505.4 |
Pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr17_+_34149820 | 0.52 |
ENSMUST00000234226.2
|
Rgl2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr17_-_56428968 | 0.43 |
ENSMUST00000041357.9
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr11_-_75313350 | 0.30 |
ENSMUST00000125982.2
ENSMUST00000137103.8 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr11_+_96941637 | 0.25 |
ENSMUST00000168565.2
|
Osbpl7
|
oxysterol binding protein-like 7 |
| chr11_-_75313412 | 0.24 |
ENSMUST00000138661.8
ENSMUST00000000769.14 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr3_-_10273628 | 0.24 |
ENSMUST00000029041.6
|
Fabp4
|
fatty acid binding protein 4, adipocyte |
| chr1_-_162687488 | 0.22 |
ENSMUST00000134098.8
ENSMUST00000111518.3 |
Fmo1
|
flavin containing monooxygenase 1 |
| chr3_-_94566107 | 0.22 |
ENSMUST00000196655.5
ENSMUST00000200407.2 ENSMUST00000006123.11 ENSMUST00000196733.5 |
Tuft1
|
tuftelin 1 |
| chr19_-_61129199 | 0.21 |
ENSMUST00000180544.3
|
Zfp950
|
zinc finger protein 950 |
| chr4_+_11558905 | 0.21 |
ENSMUST00000095145.12
ENSMUST00000108306.9 ENSMUST00000070755.13 |
Rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr1_-_88205233 | 0.20 |
ENSMUST00000065420.12
ENSMUST00000054674.15 |
Hjurp
|
Holliday junction recognition protein |
| chr11_-_8961109 | 0.20 |
ENSMUST00000020683.10
|
Hus1
|
HUS1 checkpoint clamp component |
| chr1_-_37575313 | 0.20 |
ENSMUST00000042161.15
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr17_-_28736483 | 0.19 |
ENSMUST00000114792.8
ENSMUST00000177939.8 |
Fkbp5
|
FK506 binding protein 5 |
| chr3_+_90576285 | 0.19 |
ENSMUST00000069927.10
|
S100a8
|
S100 calcium binding protein A8 (calgranulin A) |
| chr2_+_116951855 | 0.18 |
ENSMUST00000028829.13
|
Spred1
|
sprouty protein with EVH-1 domain 1, related sequence |
| chr4_-_9643636 | 0.18 |
ENSMUST00000108333.8
ENSMUST00000108334.8 ENSMUST00000108335.8 ENSMUST00000152526.8 ENSMUST00000103004.10 |
Asph
|
aspartate-beta-hydroxylase |
| chr11_+_120123727 | 0.18 |
ENSMUST00000122148.8
ENSMUST00000044985.14 |
Bahcc1
|
BAH domain and coiled-coil containing 1 |
| chr5_-_100867520 | 0.17 |
ENSMUST00000112908.2
ENSMUST00000045617.15 |
Hpse
|
heparanase |
| chr6_-_137626207 | 0.17 |
ENSMUST00000134630.6
ENSMUST00000058210.13 ENSMUST00000111878.8 |
Eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr10_+_80692948 | 0.16 |
ENSMUST00000220091.2
|
Sppl2b
|
signal peptide peptidase like 2B |
| chr4_-_123644091 | 0.16 |
ENSMUST00000102636.4
|
Akirin1
|
akirin 1 |
| chr3_-_107838895 | 0.15 |
ENSMUST00000133947.9
ENSMUST00000124215.2 ENSMUST00000106688.8 ENSMUST00000106687.9 |
Gstm7
|
glutathione S-transferase, mu 7 |
| chr1_-_88205185 | 0.15 |
ENSMUST00000147393.2
|
Hjurp
|
Holliday junction recognition protein |
| chr7_+_46496506 | 0.15 |
ENSMUST00000209984.2
|
Ldha
|
lactate dehydrogenase A |
| chr7_+_46496552 | 0.15 |
ENSMUST00000005051.6
|
Ldha
|
lactate dehydrogenase A |
| chr1_-_132953068 | 0.14 |
ENSMUST00000186617.7
ENSMUST00000067429.10 ENSMUST00000067398.13 ENSMUST00000188090.7 |
Mdm4
|
transformed mouse 3T3 cell double minute 4 |
| chr2_-_149978560 | 0.14 |
ENSMUST00000094538.6
ENSMUST00000109931.8 ENSMUST00000089207.13 |
Zfp120
|
zinc finger protein 120 |
| chr17_-_25104910 | 0.14 |
ENSMUST00000234928.2
|
Nubp2
|
nucleotide binding protein 2 |
| chr2_-_164675357 | 0.14 |
ENSMUST00000042775.5
|
Neurl2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr7_-_24997393 | 0.14 |
ENSMUST00000005583.12
|
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 |
| chr4_-_42856771 | 0.13 |
ENSMUST00000107981.2
|
Gm12394
|
predicted gene 12394 |
| chr13_+_16186410 | 0.13 |
ENSMUST00000042603.14
|
Inhba
|
inhibin beta-A |
| chr3_-_104725853 | 0.13 |
ENSMUST00000106775.8
ENSMUST00000166979.8 |
Mov10
|
Mov10 RISC complex RNA helicase |
| chr16_-_16962279 | 0.13 |
ENSMUST00000232033.2
ENSMUST00000231597.2 ENSMUST00000232540.2 |
Ccdc116
|
coiled-coil domain containing 116 |
| chr5_+_137756407 | 0.13 |
ENSMUST00000141733.8
ENSMUST00000110985.2 |
Tsc22d4
|
TSC22 domain family, member 4 |
| chr11_-_82881929 | 0.13 |
ENSMUST00000138797.2
|
Slfn9
|
schlafen 9 |
| chr11_-_82882613 | 0.13 |
ENSMUST00000092840.11
ENSMUST00000038211.13 |
Slfn9
|
schlafen 9 |
| chr17_-_25104874 | 0.13 |
ENSMUST00000234597.2
|
Nubp2
|
nucleotide binding protein 2 |
| chrX_-_100463810 | 0.13 |
ENSMUST00000118092.8
ENSMUST00000119699.8 |
Zmym3
|
zinc finger, MYM-type 3 |
| chr11_+_96941420 | 0.12 |
ENSMUST00000090020.13
|
Osbpl7
|
oxysterol binding protein-like 7 |
| chr11_+_97340962 | 0.12 |
ENSMUST00000107601.8
|
Arhgap23
|
Rho GTPase activating protein 23 |
| chr3_-_104725581 | 0.12 |
ENSMUST00000168015.8
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr16_+_18245165 | 0.12 |
ENSMUST00000206606.3
ENSMUST00000115606.9 ENSMUST00000115604.8 ENSMUST00000177856.8 ENSMUST00000178093.3 ENSMUST00000206151.3 ENSMUST00000131303.8 |
Txnrd2
|
thioredoxin reductase 2 |
| chr1_-_10108325 | 0.12 |
ENSMUST00000027050.10
ENSMUST00000188619.2 |
Cops5
|
COP9 signalosome subunit 5 |
| chr16_+_20536415 | 0.12 |
ENSMUST00000021405.8
|
Polr2h
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr11_+_96920751 | 0.12 |
ENSMUST00000021249.11
|
Scrn2
|
secernin 2 |
| chr10_+_81012465 | 0.12 |
ENSMUST00000047864.11
|
Eef2
|
eukaryotic translation elongation factor 2 |
| chr9_-_106315518 | 0.11 |
ENSMUST00000024031.13
ENSMUST00000190972.3 |
Acy1
|
aminoacylase 1 |
| chr16_+_4501934 | 0.11 |
ENSMUST00000060067.12
ENSMUST00000115854.4 ENSMUST00000229529.2 |
Dnaja3
|
DnaJ heat shock protein family (Hsp40) member A3 |
| chr11_-_97040858 | 0.11 |
ENSMUST00000118375.8
|
Tbkbp1
|
TBK1 binding protein 1 |
| chrX_+_108138965 | 0.11 |
ENSMUST00000033598.9
|
Sh3bgrl
|
SH3-binding domain glutamic acid-rich protein like |
| chr7_-_127048280 | 0.11 |
ENSMUST00000053392.11
|
Zfp689
|
zinc finger protein 689 |
| chr19_-_46917661 | 0.10 |
ENSMUST00000236727.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr7_-_24997291 | 0.10 |
ENSMUST00000148150.8
ENSMUST00000155118.2 |
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 |
| chr11_+_43572825 | 0.10 |
ENSMUST00000061070.6
ENSMUST00000094294.5 |
Pwwp2a
|
PWWP domain containing 2A |
| chr5_-_134643805 | 0.10 |
ENSMUST00000202085.2
ENSMUST00000036362.13 ENSMUST00000077636.8 |
Lat2
|
linker for activation of T cells family, member 2 |
| chr8_-_96161414 | 0.09 |
ENSMUST00000211908.2
|
Cfap20
|
cilia and flagella associated protein 20 |
| chr3_-_104725535 | 0.09 |
ENSMUST00000002297.12
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr9_-_48252176 | 0.09 |
ENSMUST00000034527.14
|
Nxpe2
|
neurexophilin and PC-esterase domain family, member 2 |
| chr10_+_118094593 | 0.09 |
ENSMUST00000219565.2
|
Gm32802
|
predicted gene, 32802 |
| chr4_+_139350152 | 0.09 |
ENSMUST00000039818.10
|
Aldh4a1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr9_-_20888054 | 0.09 |
ENSMUST00000054197.7
|
S1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr10_-_118076685 | 0.09 |
ENSMUST00000219147.2
|
Gm32717
|
predicted gene, 32717 |
| chr11_+_96822213 | 0.09 |
ENSMUST00000107633.2
|
Prr15l
|
proline rich 15-like |
| chr10_+_60925130 | 0.09 |
ENSMUST00000020298.8
|
Pcbd1
|
pterin 4 alpha carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 1 |
| chr3_+_89622323 | 0.09 |
ENSMUST00000098924.9
|
Adar
|
adenosine deaminase, RNA-specific |
| chr9_+_98178646 | 0.09 |
ENSMUST00000112938.8
ENSMUST00000112937.3 |
Nmnat3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr6_+_106746138 | 0.08 |
ENSMUST00000205163.2
|
Trnt1
|
tRNA nucleotidyl transferase, CCA-adding, 1 |
| chr17_+_32755525 | 0.08 |
ENSMUST00000169591.8
ENSMUST00000003416.15 |
Cyp4f16
|
cytochrome P450, family 4, subfamily f, polypeptide 16 |
| chr19_-_42740898 | 0.08 |
ENSMUST00000237747.2
|
Pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr9_-_107167046 | 0.08 |
ENSMUST00000035194.8
|
Mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr17_+_24689366 | 0.08 |
ENSMUST00000053024.8
|
Pgp
|
phosphoglycolate phosphatase |
| chr5_+_115061293 | 0.08 |
ENSMUST00000031540.11
ENSMUST00000112143.4 |
Oasl1
|
2'-5' oligoadenylate synthetase-like 1 |
| chr17_+_31515163 | 0.08 |
ENSMUST00000235972.2
ENSMUST00000165149.3 ENSMUST00000236251.2 |
Slc37a1
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 1 |
| chr3_+_51323383 | 0.07 |
ENSMUST00000029303.13
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr2_-_58050494 | 0.07 |
ENSMUST00000028175.7
|
Cytip
|
cytohesin 1 interacting protein |
| chr15_+_44291470 | 0.07 |
ENSMUST00000226827.2
ENSMUST00000060652.5 |
Eny2
|
ENY2 transcription and export complex 2 subunit |
| chr19_-_61129211 | 0.07 |
ENSMUST00000143264.8
ENSMUST00000205854.2 |
Zfp950
|
zinc finger protein 950 |
| chr10_+_128089965 | 0.07 |
ENSMUST00000060782.5
ENSMUST00000218722.2 |
Apon
|
apolipoprotein N |
| chr3_+_88523730 | 0.07 |
ENSMUST00000175779.8
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr10_-_18887701 | 0.07 |
ENSMUST00000105527.2
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr17_-_26138480 | 0.07 |
ENSMUST00000164982.8
ENSMUST00000179998.8 ENSMUST00000167626.8 ENSMUST00000164738.2 ENSMUST00000026826.14 ENSMUST00000167018.8 |
Rab40c
|
Rab40C, member RAS oncogene family |
| chr2_+_135501605 | 0.07 |
ENSMUST00000134310.8
|
Plcb4
|
phospholipase C, beta 4 |
| chr9_-_56068282 | 0.07 |
ENSMUST00000034876.10
|
Tspan3
|
tetraspanin 3 |
| chr3_+_129326285 | 0.07 |
ENSMUST00000197235.5
|
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr4_+_45342069 | 0.07 |
ENSMUST00000155551.8
|
Dcaf10
|
DDB1 and CUL4 associated factor 10 |
| chr4_+_117692583 | 0.07 |
ENSMUST00000169885.8
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr10_-_80513927 | 0.07 |
ENSMUST00000199949.2
|
Mknk2
|
MAP kinase-interacting serine/threonine kinase 2 |
| chr9_+_50768224 | 0.07 |
ENSMUST00000174628.8
ENSMUST00000034560.14 ENSMUST00000114437.9 ENSMUST00000175645.8 ENSMUST00000176349.8 ENSMUST00000176798.8 ENSMUST00000175640.8 |
Ppp2r1b
|
protein phosphatase 2, regulatory subunit A, beta |
| chr17_+_17594526 | 0.07 |
ENSMUST00000024620.8
|
Riok2
|
RIO kinase 2 |
| chr3_+_40755211 | 0.07 |
ENSMUST00000204473.2
|
Plk4
|
polo like kinase 4 |
| chr10_-_81127057 | 0.06 |
ENSMUST00000045744.7
|
Tjp3
|
tight junction protein 3 |
| chr5_+_114268425 | 0.06 |
ENSMUST00000031587.13
|
Ung
|
uracil DNA glycosylase |
| chr14_+_14296748 | 0.06 |
ENSMUST00000022268.10
|
Pdhb
|
pyruvate dehydrogenase (lipoamide) beta |
| chr16_+_18245134 | 0.06 |
ENSMUST00000205679.3
|
Txnrd2
|
thioredoxin reductase 2 |
| chr9_+_98178608 | 0.06 |
ENSMUST00000112935.8
|
Nmnat3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr2_+_158466802 | 0.06 |
ENSMUST00000045644.9
|
Actr5
|
ARP5 actin-related protein 5 |
| chr16_-_16962256 | 0.06 |
ENSMUST00000115711.10
|
Ccdc116
|
coiled-coil domain containing 116 |
| chr13_-_74642055 | 0.06 |
ENSMUST00000202645.4
ENSMUST00000221173.2 |
Zfp825
|
zinc finger protein 825 |
| chr2_-_126333450 | 0.06 |
ENSMUST00000040149.13
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chr11_+_116547932 | 0.06 |
ENSMUST00000116318.3
|
Prcd
|
photoreceptor disc component |
| chr7_+_127160751 | 0.06 |
ENSMUST00000190278.3
|
Tmem265
|
transmembrane protein 265 |
| chr1_+_139349912 | 0.06 |
ENSMUST00000200243.5
ENSMUST00000039867.10 |
Zbtb41
|
zinc finger and BTB domain containing 41 |
| chr1_-_152262339 | 0.06 |
ENSMUST00000162371.2
|
Tsen15
|
tRNA splicing endonuclease subunit 15 |
| chr11_-_88951511 | 0.05 |
ENSMUST00000000285.9
|
Dgke
|
diacylglycerol kinase, epsilon |
| chr13_+_49761506 | 0.05 |
ENSMUST00000021822.7
|
Ogn
|
osteoglycin |
| chr11_+_109376432 | 0.05 |
ENSMUST00000106697.8
|
Arsg
|
arylsulfatase G |
| chr4_-_149783097 | 0.05 |
ENSMUST00000038859.14
ENSMUST00000105690.9 |
Pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
| chr1_-_172034251 | 0.05 |
ENSMUST00000155109.2
|
Pea15a
|
phosphoprotein enriched in astrocytes 15A |
| chr9_-_57672124 | 0.05 |
ENSMUST00000215158.2
|
Clk3
|
CDC-like kinase 3 |
| chrX_-_101232978 | 0.05 |
ENSMUST00000033683.8
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr5_-_134343532 | 0.05 |
ENSMUST00000172715.8
ENSMUST00000174155.8 ENSMUST00000174354.8 ENSMUST00000174513.8 ENSMUST00000174772.8 ENSMUST00000173341.9 ENSMUST00000082057.10 ENSMUST00000111261.12 ENSMUST00000059042.15 |
Gtf2i
|
general transcription factor II I |
| chr15_+_81686622 | 0.05 |
ENSMUST00000109553.10
|
Tef
|
thyrotroph embryonic factor |
| chr19_-_8690330 | 0.05 |
ENSMUST00000206598.2
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr11_+_117672902 | 0.05 |
ENSMUST00000127080.9
|
Tmc8
|
transmembrane channel-like gene family 8 |
| chr10_-_127457001 | 0.05 |
ENSMUST00000049149.15
|
Lrp1
|
low density lipoprotein receptor-related protein 1 |
| chr16_+_35892437 | 0.05 |
ENSMUST00000163352.9
ENSMUST00000231468.2 |
Ccdc58
|
coiled-coil domain containing 58 |
| chr17_+_43671314 | 0.05 |
ENSMUST00000226087.2
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr17_+_32840283 | 0.05 |
ENSMUST00000077639.7
|
Cyp4f37
|
cytochrome P450, family 4, subfamily f, polypeptide 37 |
| chr9_+_77824646 | 0.05 |
ENSMUST00000034904.14
|
Elovl5
|
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
| chr12_-_79030250 | 0.05 |
ENSMUST00000070174.14
|
Tmem229b
|
transmembrane protein 229B |
| chr16_-_48814437 | 0.05 |
ENSMUST00000121869.8
|
Dzip3
|
DAZ interacting protein 3, zinc finger |
| chr1_+_71596306 | 0.05 |
ENSMUST00000027384.6
|
Atic
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr11_+_4833186 | 0.05 |
ENSMUST00000139737.2
|
Nipsnap1
|
nipsnap homolog 1 |
| chr3_-_104419128 | 0.05 |
ENSMUST00000199070.5
ENSMUST00000046316.11 ENSMUST00000198332.2 |
Lrig2
|
leucine-rich repeats and immunoglobulin-like domains 2 |
| chr11_-_117673008 | 0.04 |
ENSMUST00000152304.3
|
Tmc6
|
transmembrane channel-like gene family 6 |
| chr7_-_81439173 | 0.04 |
ENSMUST00000026092.9
|
3110040N11Rik
|
RIKEN cDNA 3110040N11 gene |
| chr2_-_114485342 | 0.04 |
ENSMUST00000055144.8
|
Dph6
|
diphthamine biosynthesis 6 |
| chr4_-_35225848 | 0.04 |
ENSMUST00000108127.4
|
C9orf72
|
C9orf72, member of C9orf72-SMCR8 complex |
| chr11_+_97689819 | 0.04 |
ENSMUST00000143571.2
|
Lasp1
|
LIM and SH3 protein 1 |
| chr2_+_84880776 | 0.04 |
ENSMUST00000111605.9
|
Tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr15_-_98729333 | 0.04 |
ENSMUST00000168846.3
|
Prkag1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr4_+_136011969 | 0.04 |
ENSMUST00000144217.8
|
Zfp46
|
zinc finger protein 46 |
| chr13_+_12580743 | 0.04 |
ENSMUST00000221560.2
ENSMUST00000071973.8 |
Ero1b
|
endoplasmic reticulum oxidoreductase 1 beta |
| chr17_-_22128907 | 0.04 |
ENSMUST00000190066.7
|
Zfp995
|
zinc finger protein 995 |
| chr5_+_34706911 | 0.04 |
ENSMUST00000118545.8
|
Sh3bp2
|
SH3-domain binding protein 2 |
| chr5_-_134343232 | 0.04 |
ENSMUST00000174867.8
|
Gtf2i
|
general transcription factor II I |
| chr8_+_69723732 | 0.04 |
ENSMUST00000212549.2
|
D130040H23Rik
|
RIKEN cDNA D130040H23 gene |
| chr12_-_112640626 | 0.04 |
ENSMUST00000001780.10
|
Akt1
|
thymoma viral proto-oncogene 1 |
| chr8_+_11777721 | 0.04 |
ENSMUST00000210104.2
|
Arhgef7
|
Rho guanine nucleotide exchange factor (GEF7) |
| chr8_-_68270936 | 0.04 |
ENSMUST00000120071.8
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr17_+_31514780 | 0.04 |
ENSMUST00000237460.2
|
Slc37a1
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 1 |
| chr1_+_172168764 | 0.04 |
ENSMUST00000056136.4
|
Kcnj10
|
potassium inwardly-rectifying channel, subfamily J, member 10 |
| chr2_-_150293406 | 0.04 |
ENSMUST00000109916.8
|
Zfp442
|
zinc finger protein 442 |
| chr3_-_63872189 | 0.04 |
ENSMUST00000029402.15
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr1_+_162398084 | 0.04 |
ENSMUST00000132158.8
ENSMUST00000135241.8 |
Vamp4
|
vesicle-associated membrane protein 4 |
| chr1_+_82817170 | 0.04 |
ENSMUST00000189220.7
ENSMUST00000113444.8 |
Agfg1
|
ArfGAP with FG repeats 1 |
| chrX_+_20529137 | 0.04 |
ENSMUST00000001989.9
|
Uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr2_-_91480096 | 0.04 |
ENSMUST00000099714.10
ENSMUST00000111333.2 |
Zfp408
|
zinc finger protein 408 |
| chr7_+_141047416 | 0.04 |
ENSMUST00000209988.2
|
Cd151
|
CD151 antigen |
| chr9_-_22218934 | 0.04 |
ENSMUST00000086278.7
ENSMUST00000215202.2 |
Zfp810
|
zinc finger protein 810 |
| chr17_+_9207180 | 0.03 |
ENSMUST00000151609.2
ENSMUST00000232775.2 ENSMUST00000136954.3 |
1700010I14Rik
|
RIKEN cDNA 1700010I14 gene |
| chr11_+_94520567 | 0.03 |
ENSMUST00000021239.7
|
Lrrc59
|
leucine rich repeat containing 59 |
| chr5_-_77262968 | 0.03 |
ENSMUST00000081964.7
|
Hopx
|
HOP homeobox |
| chr4_+_123095297 | 0.03 |
ENSMUST00000068262.6
|
Nt5c1a
|
5'-nucleotidase, cytosolic IA |
| chr9_-_78015866 | 0.03 |
ENSMUST00000162625.2
ENSMUST00000159099.8 ENSMUST00000085311.13 |
Fbxo9
|
f-box protein 9 |
| chr5_+_34706936 | 0.03 |
ENSMUST00000179943.3
|
Sh3bp2
|
SH3-domain binding protein 2 |
| chr9_-_53521585 | 0.03 |
ENSMUST00000034547.6
|
Acat1
|
acetyl-Coenzyme A acetyltransferase 1 |
| chr9_-_124075271 | 0.03 |
ENSMUST00000071300.13
ENSMUST00000185949.2 ENSMUST00000177714.8 |
2010315B03Rik
|
RIKEN cDNA 2010315B03 gene |
| chr4_-_147932817 | 0.03 |
ENSMUST00000105718.8
ENSMUST00000135798.2 |
Zfp933
|
zinc finger protein 933 |
| chr10_+_60925108 | 0.03 |
ENSMUST00000218005.2
|
Pcbd1
|
pterin 4 alpha carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 1 |
| chr7_-_30577511 | 0.03 |
ENSMUST00000214289.2
|
Cd22
|
CD22 antigen |
| chr4_+_133311665 | 0.03 |
ENSMUST00000030661.14
ENSMUST00000105899.2 |
Gpn2
|
GPN-loop GTPase 2 |
| chr3_-_63872079 | 0.03 |
ENSMUST00000161659.8
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr11_+_96920956 | 0.03 |
ENSMUST00000153482.2
|
Scrn2
|
secernin 2 |
| chr7_-_78496947 | 0.03 |
ENSMUST00000032839.13
|
Det1
|
de-etiolated homolog 1 (Arabidopsis) |
| chr7_+_26006594 | 0.03 |
ENSMUST00000098657.5
|
Cyp2a4
|
cytochrome P450, family 2, subfamily a, polypeptide 4 |
| chr7_+_141047298 | 0.03 |
ENSMUST00000106000.10
ENSMUST00000209892.2 ENSMUST00000177840.9 |
Cd151
|
CD151 antigen |
| chr1_+_172204109 | 0.03 |
ENSMUST00000052455.4
|
Pigm
|
phosphatidylinositol glycan anchor biosynthesis, class M |
| chr7_+_24981604 | 0.03 |
ENSMUST00000163320.8
ENSMUST00000005578.13 |
Cic
|
capicua transcriptional repressor |
| chr6_-_129599645 | 0.03 |
ENSMUST00000032252.8
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr12_-_112641260 | 0.03 |
ENSMUST00000144550.9
|
Akt1
|
thymoma viral proto-oncogene 1 |
| chr19_+_38384428 | 0.03 |
ENSMUST00000054098.4
|
Slc35g1
|
solute carrier family 35, member G1 |
| chr2_-_177269992 | 0.03 |
ENSMUST00000108945.8
ENSMUST00000108943.2 |
Gm14406
|
predicted gene 14406 |
| chr3_-_51184730 | 0.03 |
ENSMUST00000195432.2
ENSMUST00000091144.11 ENSMUST00000156983.3 |
Elf2
|
E74-like factor 2 |
| chr14_+_79718604 | 0.03 |
ENSMUST00000040131.13
|
Elf1
|
E74-like factor 1 |
| chr5_-_67973195 | 0.03 |
ENSMUST00000141443.2
|
Atp8a1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
| chr16_-_91728162 | 0.03 |
ENSMUST00000139277.8
ENSMUST00000154661.8 |
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr3_+_96508400 | 0.03 |
ENSMUST00000062058.5
|
Lix1l
|
Lix1-like |
| chr9_-_106764627 | 0.02 |
ENSMUST00000055843.9
|
Rbm15b
|
RNA binding motif protein 15B |
| chr2_+_122478882 | 0.02 |
ENSMUST00000142767.8
|
AA467197
|
expressed sequence AA467197 |
| chr2_+_174852034 | 0.02 |
ENSMUST00000109069.8
ENSMUST00000109070.3 |
Gm14444
|
predicted gene 14444 |
| chr2_+_132532040 | 0.02 |
ENSMUST00000148271.8
ENSMUST00000110132.3 |
Shld1
|
shieldin complex subunit 1 |
| chr4_+_59581617 | 0.02 |
ENSMUST00000107528.8
|
Hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr4_-_35225775 | 0.02 |
ENSMUST00000108126.8
|
C9orf72
|
C9orf72, member of C9orf72-SMCR8 complex |
| chr11_+_6496875 | 0.02 |
ENSMUST00000000388.15
ENSMUST00000159007.8 |
Ccm2
|
cerebral cavernous malformation 2 |
| chr1_+_51328265 | 0.02 |
ENSMUST00000051572.8
|
Cavin2
|
caveolae associated 2 |
| chr11_+_115656246 | 0.02 |
ENSMUST00000093912.11
ENSMUST00000136720.8 ENSMUST00000103034.10 ENSMUST00000141871.8 |
Tmem94
|
transmembrane protein 94 |
| chr2_-_114485421 | 0.02 |
ENSMUST00000028640.14
ENSMUST00000102542.10 |
Dph6
|
diphthamine biosynthesis 6 |
| chr17_+_17284307 | 0.02 |
ENSMUST00000127027.2
|
Zfp960
|
zinc finger protein 960 |
| chr8_-_40964282 | 0.02 |
ENSMUST00000149992.2
|
Cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr9_+_106324952 | 0.02 |
ENSMUST00000215475.2
ENSMUST00000187106.7 ENSMUST00000190167.7 |
Abhd14b
|
abhydrolase domain containing 14b |
| chr16_-_97883157 | 0.02 |
ENSMUST00000232376.2
ENSMUST00000232533.2 ENSMUST00000232346.2 |
A630089N07Rik
|
RIKEN cDNA A630089N07 gene |
| chr16_+_48814548 | 0.02 |
ENSMUST00000117994.8
ENSMUST00000048374.6 |
Cip2a
|
cell proliferation regulating inhibitor of protein phosphatase 2A |
| chr18_-_61344644 | 0.02 |
ENSMUST00000146409.8
|
Slc26a2
|
solute carrier family 26 (sulfate transporter), member 2 |
| chr8_+_11778039 | 0.02 |
ENSMUST00000110909.9
|
Arhgef7
|
Rho guanine nucleotide exchange factor (GEF7) |
| chr9_-_106324642 | 0.02 |
ENSMUST00000185334.7
ENSMUST00000187001.2 ENSMUST00000171678.9 ENSMUST00000190798.7 ENSMUST00000048685.13 ENSMUST00000171925.8 |
Abhd14a
|
abhydrolase domain containing 14A |
| chr6_+_48570817 | 0.02 |
ENSMUST00000154010.8
ENSMUST00000009420.15 ENSMUST00000163452.7 ENSMUST00000118229.2 ENSMUST00000135151.3 |
Repin1
|
replication initiator 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tlx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.4 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.2 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.1 | 0.5 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.2 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 0.0 | 0.5 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.5 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.3 | GO:0019661 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.1 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.1 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 0.2 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.1 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0034147 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.0 | 0.1 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.2 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.1 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.2 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.0 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.3 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.0 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.0 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:0060356 | leucine import(GO:0060356) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.5 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.0 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.0 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.0 | 0.1 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.4 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |