Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
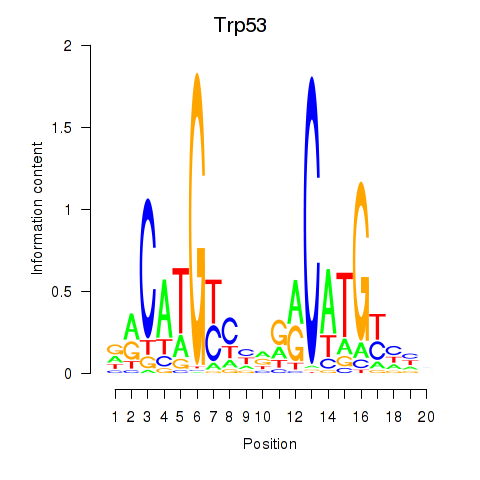
Results for Trp53
Z-value: 1.08
Transcription factors associated with Trp53
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Trp53
|
ENSMUSG00000059552.14 | transformation related protein 53 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Trp53 | mm39_v1_chr11_+_69471185_69471208 | 1.00 | 7.9e-05 | Click! |
Activity profile of Trp53 motif
Sorted Z-values of Trp53 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_22220000 | 1.61 |
ENSMUST00000110455.4
|
H2bc12
|
H2B clustered histone 12 |
| chr13_+_21938258 | 1.41 |
ENSMUST00000091709.3
|
H2bc15
|
H2B clustered histone 15 |
| chr13_-_21994366 | 1.31 |
ENSMUST00000091749.4
|
H2bc23
|
H2B clustered histone 23 |
| chr13_-_22219738 | 1.18 |
ENSMUST00000091742.6
|
H2ac12
|
H2A clustered histone 12 |
| chr13_+_22227359 | 1.03 |
ENSMUST00000110452.2
|
H2bc11
|
H2B clustered histone 11 |
| chr5_-_66238313 | 0.77 |
ENSMUST00000202700.4
ENSMUST00000094757.9 ENSMUST00000113724.6 |
Rbm47
|
RNA binding motif protein 47 |
| chr8_-_69541852 | 0.77 |
ENSMUST00000037478.13
ENSMUST00000148856.2 |
Slc18a1
|
solute carrier family 18 (vesicular monoamine), member 1 |
| chr1_-_73967664 | 0.73 |
ENSMUST00000187691.7
|
Tns1
|
tensin 1 |
| chr7_+_24583994 | 0.69 |
ENSMUST00000108428.8
|
Rps19
|
ribosomal protein S19 |
| chr13_-_98951890 | 0.68 |
ENSMUST00000040340.16
ENSMUST00000179563.8 ENSMUST00000109403.2 |
Fcho2
|
FCH domain only 2 |
| chr7_-_3828640 | 0.66 |
ENSMUST00000189095.7
ENSMUST00000094911.5 ENSMUST00000153846.8 ENSMUST00000108619.8 ENSMUST00000108620.8 |
Gm15448
|
predicted gene 15448 |
| chr13_-_23755374 | 0.65 |
ENSMUST00000102969.6
|
H2ac8
|
H2A clustered histone 8 |
| chr6_-_38814159 | 0.62 |
ENSMUST00000160962.8
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr13_-_22808421 | 0.62 |
ENSMUST00000227244.2
|
Vmn1r206
|
vomeronasal 1 receptor 206 |
| chr10_-_128245501 | 0.61 |
ENSMUST00000172348.8
ENSMUST00000166608.8 ENSMUST00000164199.8 ENSMUST00000171370.2 ENSMUST00000026439.14 |
Nabp2
|
nucleic acid binding protein 2 |
| chr14_-_70412804 | 0.59 |
ENSMUST00000143393.2
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr13_+_23758555 | 0.56 |
ENSMUST00000090776.7
|
H2ac7
|
H2A clustered histone 7 |
| chr3_-_75864195 | 0.50 |
ENSMUST00000038563.14
ENSMUST00000167078.8 ENSMUST00000117242.8 |
Golim4
|
golgi integral membrane protein 4 |
| chr4_-_118795809 | 0.50 |
ENSMUST00000215312.3
|
Olfr1328
|
olfactory receptor 1328 |
| chr9_+_106247943 | 0.48 |
ENSMUST00000173748.2
|
Dusp7
|
dual specificity phosphatase 7 |
| chr13_-_22913799 | 0.47 |
ENSMUST00000237024.2
ENSMUST00000236800.2 |
Vmn1r207
|
vomeronasal 1 receptor 207 |
| chr11_+_58839716 | 0.46 |
ENSMUST00000078267.5
|
H2bu2
|
H2B.U histone 2 |
| chr13_-_22227114 | 0.46 |
ENSMUST00000091741.6
|
H2ac11
|
H2A clustered histone 11 |
| chr5_+_52521133 | 0.45 |
ENSMUST00000101208.6
|
Sod3
|
superoxide dismutase 3, extracellular |
| chr14_-_104081119 | 0.44 |
ENSMUST00000227824.2
ENSMUST00000172237.2 |
Ednrb
|
endothelin receptor type B |
| chr10_+_110581293 | 0.44 |
ENSMUST00000174857.8
ENSMUST00000073781.12 ENSMUST00000173471.8 ENSMUST00000173634.2 |
E2f7
|
E2F transcription factor 7 |
| chr13_-_22993852 | 0.43 |
ENSMUST00000227038.2
ENSMUST00000227265.2 |
Vmn1r209
|
vomeronasal 1 receptor 209 |
| chr9_+_72892693 | 0.42 |
ENSMUST00000037977.15
|
Ccpg1
|
cell cycle progression 1 |
| chr7_-_24459736 | 0.42 |
ENSMUST00000063956.7
|
Cd177
|
CD177 antigen |
| chr15_-_102154874 | 0.42 |
ENSMUST00000063339.14
|
Rarg
|
retinoic acid receptor, gamma |
| chr13_-_27766147 | 0.40 |
ENSMUST00000006664.8
ENSMUST00000095926.2 |
Prl8a1
|
prolactin family 8, subfamily a, member 1 |
| chr2_+_29779227 | 0.40 |
ENSMUST00000123335.8
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr7_-_103710652 | 0.39 |
ENSMUST00000074064.5
|
4930516K23Rik
|
RIKEN cDNA 4930516K23 gene |
| chr15_-_76501525 | 0.38 |
ENSMUST00000230977.2
|
Slc39a4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr19_+_44191704 | 0.37 |
ENSMUST00000026220.7
|
Scd3
|
stearoyl-coenzyme A desaturase 3 |
| chr3_+_105821450 | 0.37 |
ENSMUST00000198080.5
ENSMUST00000199977.2 |
Tmigd3
|
transmembrane and immunoglobulin domain containing 3 |
| chr13_-_74956924 | 0.36 |
ENSMUST00000223206.2
|
Cast
|
calpastatin |
| chr2_+_37080286 | 0.36 |
ENSMUST00000218602.2
|
Olfr365
|
olfactory receptor 365 |
| chr9_+_72892850 | 0.35 |
ENSMUST00000150826.9
ENSMUST00000085350.11 ENSMUST00000140675.8 |
Ccpg1
|
cell cycle progression 1 |
| chr4_+_111830119 | 0.35 |
ENSMUST00000106568.8
ENSMUST00000055014.11 ENSMUST00000163281.2 |
Skint7
|
selection and upkeep of intraepithelial T cells 7 |
| chr11_-_79037213 | 0.34 |
ENSMUST00000018478.11
ENSMUST00000108264.8 |
Ksr1
|
kinase suppressor of ras 1 |
| chr10_+_58649181 | 0.34 |
ENSMUST00000135526.9
ENSMUST00000153031.2 |
Sh3rf3
|
SH3 domain containing ring finger 3 |
| chr7_-_140087224 | 0.33 |
ENSMUST00000209873.2
ENSMUST00000064392.8 ENSMUST00000215768.2 ENSMUST00000215340.2 |
Olfr536
|
olfactory receptor 536 |
| chr5_-_138270995 | 0.31 |
ENSMUST00000161665.2
ENSMUST00000100530.8 ENSMUST00000161279.8 ENSMUST00000161647.8 |
Gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr1_-_88133472 | 0.30 |
ENSMUST00000119972.4
|
Dnajb3
|
DnaJ heat shock protein family (Hsp40) member B3 |
| chr13_-_74956640 | 0.30 |
ENSMUST00000231578.2
|
Cast
|
calpastatin |
| chr11_-_65636651 | 0.30 |
ENSMUST00000138093.2
|
Map2k4
|
mitogen-activated protein kinase kinase 4 |
| chr6_-_131224305 | 0.27 |
ENSMUST00000032306.15
ENSMUST00000088867.7 |
Klra2
|
killer cell lectin-like receptor, subfamily A, member 2 |
| chrX_-_37723943 | 0.27 |
ENSMUST00000058265.8
|
C1galt1c1
|
C1GALT1-specific chaperone 1 |
| chr3_-_57599956 | 0.27 |
ENSMUST00000238789.2
ENSMUST00000197088.5 ENSMUST00000099091.4 |
Ankub1
|
ankrin repeat and ubiquitin domain containing 1 |
| chr1_-_184615415 | 0.27 |
ENSMUST00000048308.6
|
C130074G19Rik
|
RIKEN cDNA C130074G19 gene |
| chr17_+_47747657 | 0.27 |
ENSMUST00000150819.3
|
AI661453
|
expressed sequence AI661453 |
| chr9_-_114811807 | 0.27 |
ENSMUST00000053150.8
|
Rps27rt
|
ribosomal protein S27, retrogene |
| chr7_-_96981221 | 0.26 |
ENSMUST00000139582.9
|
Usp35
|
ubiquitin specific peptidase 35 |
| chr3_+_63988968 | 0.26 |
ENSMUST00000029406.6
|
Vmn2r1
|
vomeronasal 2, receptor 1 |
| chr16_+_38279289 | 0.26 |
ENSMUST00000099816.3
ENSMUST00000232409.2 |
Cd80
|
CD80 antigen |
| chr9_+_107454114 | 0.25 |
ENSMUST00000112387.9
ENSMUST00000123005.8 ENSMUST00000010195.14 ENSMUST00000144392.2 |
Hyal1
|
hyaluronoglucosaminidase 1 |
| chr5_-_23889607 | 0.25 |
ENSMUST00000197985.5
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr10_+_56253418 | 0.24 |
ENSMUST00000068581.9
ENSMUST00000217789.2 |
Gja1
|
gap junction protein, alpha 1 |
| chr8_+_124133832 | 0.24 |
ENSMUST00000098324.4
|
Mc1r
|
melanocortin 1 receptor |
| chr3_+_20043315 | 0.23 |
ENSMUST00000173779.2
|
Cp
|
ceruloplasmin |
| chrX_+_73298388 | 0.23 |
ENSMUST00000119197.8
ENSMUST00000088313.5 |
Emd
|
emerin |
| chr17_+_32725420 | 0.22 |
ENSMUST00000235238.2
ENSMUST00000165999.2 |
Cyp4f17
|
cytochrome P450, family 4, subfamily f, polypeptide 17 |
| chr7_+_75105282 | 0.21 |
ENSMUST00000207750.2
ENSMUST00000166315.7 |
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr11_-_98329782 | 0.21 |
ENSMUST00000002655.8
|
Mien1
|
migration and invasion enhancer 1 |
| chr19_+_4905158 | 0.21 |
ENSMUST00000119694.3
ENSMUST00000237504.2 ENSMUST00000237011.2 |
Ctsf
|
cathepsin F |
| chr13_-_23372145 | 0.21 |
ENSMUST00000228239.2
ENSMUST00000227950.2 ENSMUST00000226651.2 ENSMUST00000227679.2 ENSMUST00000228854.2 |
Vmn1r220
|
vomeronasal 1 receptor 220 |
| chr9_+_38202403 | 0.21 |
ENSMUST00000216276.2
ENSMUST00000215219.2 |
Olfr896-ps1
|
olfactory receptor 896, pseudogene 1 |
| chr9_+_72892786 | 0.20 |
ENSMUST00000156879.8
|
Ccpg1
|
cell cycle progression 1 |
| chr7_+_5037117 | 0.18 |
ENSMUST00000076791.4
|
4632433K11Rik
|
RIKEN cDNA 4632433K11 gene |
| chrX_+_135723420 | 0.18 |
ENSMUST00000033800.13
|
Plp1
|
proteolipid protein (myelin) 1 |
| chr3_+_59832635 | 0.18 |
ENSMUST00000049476.3
|
Aadacl2fm1
|
AADACL2 family member 1 |
| chr12_+_59142439 | 0.17 |
ENSMUST00000219140.3
|
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr17_-_47748289 | 0.17 |
ENSMUST00000061885.9
|
1700001C19Rik
|
RIKEN cDNA 1700001C19 gene |
| chr2_+_136733421 | 0.17 |
ENSMUST00000141463.8
|
Slx4ip
|
SLX4 interacting protein |
| chr17_-_10538253 | 0.17 |
ENSMUST00000233828.2
ENSMUST00000233645.2 ENSMUST00000042296.9 |
Qk
|
quaking, KH domain containing RNA binding |
| chr8_+_95703506 | 0.17 |
ENSMUST00000212581.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr1_+_88030951 | 0.17 |
ENSMUST00000113135.6
ENSMUST00000113138.8 |
Ugt1a7c
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A7C UDP glucuronosyltransferase 1 family, polypeptide A6B |
| chr19_-_4843860 | 0.16 |
ENSMUST00000178615.8
ENSMUST00000179189.9 ENSMUST00000237760.2 ENSMUST00000164376.3 ENSMUST00000164209.9 ENSMUST00000180248.8 |
Rbm4
|
RNA binding motif protein 4 |
| chr4_+_148642879 | 0.16 |
ENSMUST00000017408.14
ENSMUST00000076022.7 |
Exosc10
|
exosome component 10 |
| chr5_-_135494775 | 0.16 |
ENSMUST00000212301.2
|
Hip1
|
huntingtin interacting protein 1 |
| chr14_-_73563212 | 0.15 |
ENSMUST00000022701.7
|
Rb1
|
RB transcriptional corepressor 1 |
| chr2_-_17735847 | 0.15 |
ENSMUST00000028080.12
|
Nebl
|
nebulette |
| chr2_+_72115981 | 0.15 |
ENSMUST00000090824.12
ENSMUST00000135469.8 |
Map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr11_-_102815910 | 0.15 |
ENSMUST00000021311.10
|
Kif18b
|
kinesin family member 18B |
| chr1_+_172327569 | 0.14 |
ENSMUST00000111230.8
|
Tagln2
|
transgelin 2 |
| chr7_+_96981517 | 0.14 |
ENSMUST00000054107.6
|
Kctd21
|
potassium channel tetramerisation domain containing 21 |
| chr11_+_78427181 | 0.14 |
ENSMUST00000128788.8
|
Ift20
|
intraflagellar transport 20 |
| chr7_+_24584076 | 0.14 |
ENSMUST00000153451.9
ENSMUST00000108429.8 |
Rps19
|
ribosomal protein S19 |
| chr9_+_61280501 | 0.14 |
ENSMUST00000162583.8
ENSMUST00000161993.8 ENSMUST00000160882.8 ENSMUST00000160724.8 ENSMUST00000162973.8 ENSMUST00000159050.8 |
Tle3
|
transducin-like enhancer of split 3 |
| chr6_-_136918671 | 0.14 |
ENSMUST00000032344.12
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr2_-_132787790 | 0.14 |
ENSMUST00000038280.5
|
Fermt1
|
fermitin family member 1 |
| chr19_-_5560473 | 0.13 |
ENSMUST00000237111.2
ENSMUST00000070172.6 |
Snx32
|
sorting nexin 32 |
| chr9_+_78303059 | 0.13 |
ENSMUST00000113367.2
|
Ddx43
|
DEAD box helicase 43 |
| chr7_+_64042482 | 0.13 |
ENSMUST00000206882.2
|
Mcee
|
methylmalonyl CoA epimerase |
| chr11_+_66802807 | 0.13 |
ENSMUST00000123434.3
|
Pirt
|
phosphoinositide-interacting regulator of transient receptor potential channels |
| chr2_-_150204628 | 0.13 |
ENSMUST00000051153.6
|
3300002I08Rik
|
RIKEN cDNA 3300002I08 gene |
| chr16_-_11727262 | 0.13 |
ENSMUST00000127972.8
ENSMUST00000121750.2 ENSMUST00000096272.11 ENSMUST00000073371.7 |
Cpped1
|
calcineurin-like phosphoesterase domain containing 1 |
| chr2_+_37029334 | 0.12 |
ENSMUST00000214905.2
ENSMUST00000217298.2 ENSMUST00000104995.3 |
Olfr364-ps1
|
olfactory receptor 364, pseudogene 1 |
| chr1_-_38704028 | 0.12 |
ENSMUST00000039827.14
|
Aff3
|
AF4/FMR2 family, member 3 |
| chr14_+_26836803 | 0.11 |
ENSMUST00000225949.2
|
Arhgef3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr4_+_143785696 | 0.11 |
ENSMUST00000091676.12
ENSMUST00000133626.8 |
Pramel4
|
PRAME like 4 |
| chr7_+_24099346 | 0.11 |
ENSMUST00000108434.2
|
Smg9
|
smg-9 homolog, nonsense mediated mRNA decay factor (C. elegans) |
| chr4_-_135300934 | 0.11 |
ENSMUST00000105855.2
|
Grhl3
|
grainyhead like transcription factor 3 |
| chr1_-_160958998 | 0.11 |
ENSMUST00000111611.8
|
Klhl20
|
kelch-like 20 |
| chr7_-_119461027 | 0.10 |
ENSMUST00000137888.2
ENSMUST00000142120.2 |
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr6_-_136918885 | 0.10 |
ENSMUST00000111891.4
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chrX_+_73298285 | 0.10 |
ENSMUST00000002029.13
|
Emd
|
emerin |
| chr11_+_73158214 | 0.10 |
ENSMUST00000049676.3
|
Trpv3
|
transient receptor potential cation channel, subfamily V, member 3 |
| chr12_+_59176506 | 0.10 |
ENSMUST00000175912.8
ENSMUST00000176892.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr12_-_91351177 | 0.10 |
ENSMUST00000141429.8
|
Cep128
|
centrosomal protein 128 |
| chr14_+_30673334 | 0.09 |
ENSMUST00000226551.2
ENSMUST00000228328.2 |
Nek4
|
NIMA (never in mitosis gene a)-related expressed kinase 4 |
| chr6_-_136918495 | 0.09 |
ENSMUST00000111892.8
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr3_+_116653113 | 0.09 |
ENSMUST00000040260.11
|
Frrs1
|
ferric-chelate reductase 1 |
| chr1_+_133278248 | 0.09 |
ENSMUST00000094556.3
|
Ren1
|
renin 1 structural |
| chr2_+_148237258 | 0.09 |
ENSMUST00000109962.4
|
Sstr4
|
somatostatin receptor 4 |
| chr6_-_116438116 | 0.09 |
ENSMUST00000026795.13
|
Alox5
|
arachidonate 5-lipoxygenase |
| chr6_-_124698805 | 0.09 |
ENSMUST00000173315.8
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr11_-_99447672 | 0.08 |
ENSMUST00000092699.3
|
Krtap3-2
|
keratin associated protein 3-2 |
| chr7_+_24099037 | 0.08 |
ENSMUST00000002280.11
|
Smg9
|
smg-9 homolog, nonsense mediated mRNA decay factor (C. elegans) |
| chr19_-_12901783 | 0.08 |
ENSMUST00000213713.2
ENSMUST00000216888.2 ENSMUST00000213177.2 |
Olfr1448
|
olfactory receptor 1448 |
| chr17_+_47747540 | 0.07 |
ENSMUST00000037701.13
|
AI661453
|
expressed sequence AI661453 |
| chr6_-_118539187 | 0.07 |
ENSMUST00000112830.3
|
Ankrd26
|
ankyrin repeat domain 26 |
| chr18_-_35836161 | 0.07 |
ENSMUST00000025208.7
|
Dnajc18
|
DnaJ heat shock protein family (Hsp40) member C18 |
| chr9_-_70048766 | 0.07 |
ENSMUST00000034749.16
|
Fam81a
|
family with sequence similarity 81, member A |
| chr6_+_67586695 | 0.07 |
ENSMUST00000103303.3
|
Igkv1-135
|
immunoglobulin kappa variable 1-135 |
| chr11_+_78427219 | 0.07 |
ENSMUST00000050366.15
ENSMUST00000108275.2 |
Ift20
|
intraflagellar transport 20 |
| chr16_-_94023976 | 0.07 |
ENSMUST00000227698.2
|
Hlcs
|
holocarboxylase synthetase (biotin- [propriony-Coenzyme A-carboxylase (ATP-hydrolysing)] ligase) |
| chr7_-_64522741 | 0.06 |
ENSMUST00000094331.5
|
Nsmce3
|
NSE3 homolog, SMC5-SMC6 complex component |
| chr9_+_65268304 | 0.06 |
ENSMUST00000147185.3
|
Ubap1l
|
ubiquitin-associated protein 1-like |
| chr12_+_59176543 | 0.06 |
ENSMUST00000069430.15
ENSMUST00000177370.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr8_-_22675773 | 0.06 |
ENSMUST00000046916.9
|
Ckap2
|
cytoskeleton associated protein 2 |
| chr6_+_39569502 | 0.06 |
ENSMUST00000135671.8
ENSMUST00000119379.2 |
Ndufb2
|
NADH:ubiquinone oxidoreductase subunit B2 |
| chr4_+_116414855 | 0.05 |
ENSMUST00000030460.15
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr7_+_16042255 | 0.05 |
ENSMUST00000209688.2
|
Bbc3
|
BCL2 binding component 3 |
| chr10_-_81186137 | 0.05 |
ENSMUST00000167481.8
|
Hmg20b
|
high mobility group 20B |
| chr7_-_103390529 | 0.05 |
ENSMUST00000051346.5
|
Olfr629
|
olfactory receptor 629 |
| chr14_+_73475335 | 0.05 |
ENSMUST00000044405.8
|
Lpar6
|
lysophosphatidic acid receptor 6 |
| chr18_+_12874390 | 0.04 |
ENSMUST00000121018.8
ENSMUST00000119108.8 ENSMUST00000186263.2 ENSMUST00000191078.7 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chr6_+_68414401 | 0.04 |
ENSMUST00000103324.3
|
Igkv15-103
|
immunoglobulin kappa chain variable 15-103 |
| chr8_-_89362745 | 0.04 |
ENSMUST00000034087.9
|
Snx20
|
sorting nexin 20 |
| chr12_-_78953703 | 0.04 |
ENSMUST00000021544.8
|
Plek2
|
pleckstrin 2 |
| chr15_-_98796373 | 0.04 |
ENSMUST00000229775.2
ENSMUST00000023737.6 |
Dhh
|
desert hedgehog |
| chr14_-_70890521 | 0.04 |
ENSMUST00000062629.5
|
Npm2
|
nucleophosmin/nucleoplasmin 2 |
| chr19_-_47126719 | 0.04 |
ENSMUST00000140512.8
ENSMUST00000035822.2 |
Calhm2
|
calcium homeostasis modulator family member 2 |
| chr1_-_22386016 | 0.04 |
ENSMUST00000164877.8
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chrX_+_73298342 | 0.04 |
ENSMUST00000096424.11
|
Emd
|
emerin |
| chr12_+_117497478 | 0.04 |
ENSMUST00000220781.2
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr4_-_135112956 | 0.04 |
ENSMUST00000105857.8
ENSMUST00000105858.8 ENSMUST00000064481.15 ENSMUST00000123632.2 |
Ncmap
|
noncompact myelin associated protein |
| chr6_-_128868068 | 0.04 |
ENSMUST00000178918.2
ENSMUST00000160290.8 |
BC035044
|
cDNA sequence BC035044 |
| chr14_-_119162736 | 0.03 |
ENSMUST00000004055.10
|
Dzip1
|
DAZ interacting protein 1 |
| chr11_-_73290321 | 0.03 |
ENSMUST00000131253.2
ENSMUST00000120303.9 |
Olfr1
|
olfactory receptor 1 |
| chr8_-_91860655 | 0.03 |
ENSMUST00000125257.3
|
Aktip
|
AKT interacting protein |
| chr11_-_8989582 | 0.03 |
ENSMUST00000043377.6
|
Sun3
|
Sad1 and UNC84 domain containing 3 |
| chr11_+_116484513 | 0.03 |
ENSMUST00000153476.8
|
Aanat
|
arylalkylamine N-acetyltransferase |
| chr1_-_74343279 | 0.03 |
ENSMUST00000141560.2
|
Tmbim1
|
transmembrane BAX inhibitor motif containing 1 |
| chr4_-_118774662 | 0.03 |
ENSMUST00000071979.2
|
Olfr1329
|
olfactory receptor 1329 |
| chr9_+_32305259 | 0.03 |
ENSMUST00000172015.3
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr11_-_32217547 | 0.02 |
ENSMUST00000109389.9
ENSMUST00000129010.2 ENSMUST00000020530.12 |
Nprl3
|
nitrogen permease regulator-like 3 |
| chr9_-_58065800 | 0.02 |
ENSMUST00000168864.4
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr11_-_115968373 | 0.02 |
ENSMUST00000174822.8
|
Unc13d
|
unc-13 homolog D |
| chr8_-_65471175 | 0.02 |
ENSMUST00000078409.5
ENSMUST00000048565.9 |
Trim61
Trim60
|
tripartite motif-containing 61 tripartite motif-containing 60 |
| chr2_-_89084406 | 0.02 |
ENSMUST00000215447.2
|
Olfr1228
|
olfactory receptor 1228 |
| chr5_-_143166476 | 0.02 |
ENSMUST00000049861.11
ENSMUST00000165318.4 |
Rbak
|
RB-associated KRAB zinc finger |
| chr16_-_94657531 | 0.02 |
ENSMUST00000232562.2
ENSMUST00000165538.3 |
Kcnj6
|
potassium inwardly-rectifying channel, subfamily J, member 6 |
| chr4_-_144007036 | 0.02 |
ENSMUST00000035757.13
|
Pramel5
|
PRAME like 5 |
| chr9_-_19968858 | 0.02 |
ENSMUST00000216538.2
ENSMUST00000212098.3 |
Olfr867
|
olfactory receptor 867 |
| chr13_+_59860096 | 0.02 |
ENSMUST00000165133.3
|
Spata31d1b
|
spermatogenesis associated 31 subfamily D, member 1B |
| chr2_-_122144125 | 0.02 |
ENSMUST00000147788.8
ENSMUST00000154412.2 ENSMUST00000110537.8 ENSMUST00000148417.8 |
Duoxa1
|
dual oxidase maturation factor 1 |
| chr5_+_143166759 | 0.01 |
ENSMUST00000031574.10
|
Spdye4b
|
speedy/RINGO cell cycle regulator family, member E4B |
| chr6_+_71684846 | 0.01 |
ENSMUST00000212792.2
|
Reep1
|
receptor accessory protein 1 |
| chr4_+_43983472 | 0.01 |
ENSMUST00000095107.3
|
Ccin
|
calicin |
| chr10_-_81186025 | 0.01 |
ENSMUST00000122993.8
|
Hmg20b
|
high mobility group 20B |
| chr7_-_18301211 | 0.01 |
ENSMUST00000094794.5
|
Psg27
|
pregnancy-specific glycoprotein 27 |
| chr8_+_84748073 | 0.01 |
ENSMUST00000055077.7
|
Palm3
|
paralemmin 3 |
| chr14_+_55998847 | 0.01 |
ENSMUST00000044554.5
|
Ltb4r2
|
leukotriene B4 receptor 2 |
| chr7_+_19585919 | 0.01 |
ENSMUST00000172815.4
|
Gm19345
|
predicted gene, 19345 |
| chr5_+_138019523 | 0.01 |
ENSMUST00000171498.3
ENSMUST00000085886.3 |
Smok3a
|
sperm motility kinase 3A |
| chr11_-_7138569 | 0.01 |
ENSMUST00000122910.3
|
Ccdc201
|
coiled coil domain 201 |
| chr11_+_81948649 | 0.01 |
ENSMUST00000000342.3
|
Ccl11
|
chemokine (C-C motif) ligand 11 |
| chr5_+_138035486 | 0.01 |
ENSMUST00000100537.3
|
Smok3b
|
sperm motility kinase 3B |
| chr5_+_144127102 | 0.01 |
ENSMUST00000060747.8
|
Bhlha15
|
basic helix-loop-helix family, member a15 |
| chr4_+_114914607 | 0.01 |
ENSMUST00000136946.8
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr9_+_26910995 | 0.01 |
ENSMUST00000213770.2
ENSMUST00000213683.2 ENSMUST00000039161.10 |
Thyn1
|
thymocyte nuclear protein 1 |
| chr5_+_138051456 | 0.01 |
ENSMUST00000110967.4
|
Smok3c
|
sperm motility kinase 3C |
| chr11_+_77928736 | 0.01 |
ENSMUST00000072289.12
ENSMUST00000100784.9 ENSMUST00000073660.7 |
Flot2
|
flotillin 2 |
| chr9_+_45313913 | 0.01 |
ENSMUST00000214257.2
|
Fxyd2
|
FXYD domain-containing ion transport regulator 2 |
| chr19_-_5399368 | 0.01 |
ENSMUST00000238111.2
|
Cst6
|
cystatin E/M |
| chr1_+_82817794 | 0.01 |
ENSMUST00000186043.2
|
Agfg1
|
ArfGAP with FG repeats 1 |
| chr2_+_86655007 | 0.01 |
ENSMUST00000217509.2
|
Olfr1094
|
olfactory receptor 1094 |
| chr7_-_24245419 | 0.01 |
ENSMUST00000011776.8
|
Pinlyp
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr3_-_93600377 | 0.01 |
ENSMUST00000193565.2
|
Tdpoz6
|
TD and POZ domain containing 6 |
| chr7_+_102834996 | 0.01 |
ENSMUST00000218618.2
|
Olfr592
|
olfactory receptor 592 |
| chr2_-_160169414 | 0.01 |
ENSMUST00000099127.3
|
Gm826
|
predicted gene 826 |
| chr1_+_75377616 | 0.01 |
ENSMUST00000122266.3
|
Speg
|
SPEG complex locus |
| chr1_+_40364752 | 0.01 |
ENSMUST00000193388.2
|
Il1rl2
|
interleukin 1 receptor-like 2 |
| chr18_+_84970235 | 0.01 |
ENSMUST00000223789.3
|
Fbxo15
|
F-box protein 15 |
| chr7_+_18451998 | 0.01 |
ENSMUST00000051973.9
ENSMUST00000208221.2 ENSMUST00000108481.8 |
Psg22
|
pregnancy-specific glycoprotein 22 |
| chr5_-_116560916 | 0.01 |
ENSMUST00000036991.5
|
Hspb8
|
heat shock protein 8 |
| chr2_-_89084074 | 0.01 |
ENSMUST00000216392.2
|
Olfr1228
|
olfactory receptor 1228 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Trp53
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.1 | 0.6 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 | 1.0 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.8 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 0.4 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 | 0.2 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.1 | 0.2 | GO:0003294 | atrial ventricular junction remodeling(GO:0003294) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.1 | 0.4 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.1 | 0.4 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.1 | 0.6 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.4 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.2 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.0 | 0.1 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 0.0 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.2 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.7 | GO:0007343 | egg activation(GO:0007343) negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:1900062 | regulation of replicative cell aging(GO:1900062) |
| 0.0 | 0.3 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.3 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.2 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.2 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.6 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.0 | 0.1 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.7 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.4 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0070781 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 0.0 | 0.1 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.0 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.1 | 0.6 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:0098890 | extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.2 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.4 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.0 | 0.1 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.2 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0005922 | fascia adherens(GO:0005916) connexon complex(GO:0005922) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.4 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 0.7 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.2 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.4 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.4 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 0.3 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.3 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.2 | GO:1903763 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.6 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.6 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.2 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.3 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0004080 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |