Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
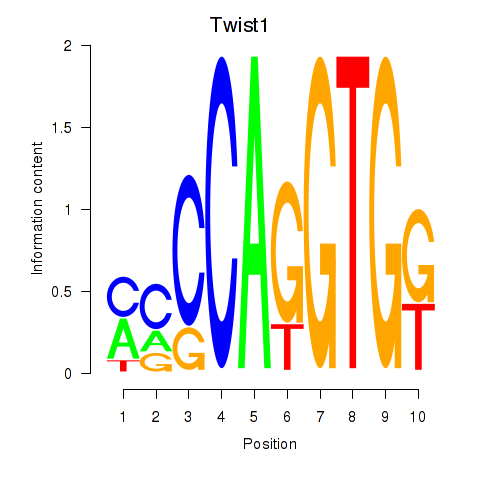
Results for Twist1
Z-value: 0.35
Transcription factors associated with Twist1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Twist1
|
ENSMUSG00000035799.7 | twist basic helix-loop-helix transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Twist1 | mm39_v1_chr12_+_34007645_34007670 | 0.02 | 9.7e-01 | Click! |
Activity profile of Twist1 motif
Sorted Z-values of Twist1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_126866682 | 0.29 |
ENSMUST00000040560.11
|
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr6_-_124790029 | 0.22 |
ENSMUST00000149610.3
|
Tpi1
|
triosephosphate isomerase 1 |
| chr9_-_44253630 | 0.21 |
ENSMUST00000097558.5
|
Hmbs
|
hydroxymethylbilane synthase |
| chr7_+_82516491 | 0.20 |
ENSMUST00000082237.7
|
Mex3b
|
mex3 RNA binding family member B |
| chr6_-_124410452 | 0.19 |
ENSMUST00000124998.2
ENSMUST00000238807.2 |
Clstn3
|
calsyntenin 3 |
| chr11_+_53661251 | 0.18 |
ENSMUST00000138913.8
ENSMUST00000123376.8 ENSMUST00000019043.13 ENSMUST00000133291.3 |
Irf1
|
interferon regulatory factor 1 |
| chr10_+_61010983 | 0.17 |
ENSMUST00000143207.8
ENSMUST00000148181.8 ENSMUST00000151886.8 |
Tbata
|
thymus, brain and testes associated |
| chr17_+_34029484 | 0.15 |
ENSMUST00000048560.11
ENSMUST00000172649.8 ENSMUST00000173789.2 |
Kank3
|
KN motif and ankyrin repeat domains 3 |
| chr17_+_74645936 | 0.15 |
ENSMUST00000224711.2
ENSMUST00000024869.8 ENSMUST00000233611.2 |
Spast
|
spastin |
| chrX_-_100310959 | 0.14 |
ENSMUST00000135038.2
|
Il2rg
|
interleukin 2 receptor, gamma chain |
| chr7_-_126817475 | 0.13 |
ENSMUST00000106313.8
ENSMUST00000142356.3 |
Septin1
|
septin 1 |
| chr7_-_30814652 | 0.12 |
ENSMUST00000168884.8
ENSMUST00000108102.9 |
Hpn
|
hepsin |
| chr15_+_100251030 | 0.12 |
ENSMUST00000075675.7
ENSMUST00000088142.6 ENSMUST00000176287.2 |
Methig1
Mettl7a2
|
methyltransferase hypoxia inducible domain containing 1 methyltransferase like 7A2 |
| chr15_-_89007290 | 0.12 |
ENSMUST00000109353.9
|
Tubgcp6
|
tubulin, gamma complex associated protein 6 |
| chr4_+_132495636 | 0.12 |
ENSMUST00000102561.11
|
Rpa2
|
replication protein A2 |
| chr8_-_71834543 | 0.11 |
ENSMUST00000002466.9
|
Nr2f6
|
nuclear receptor subfamily 2, group F, member 6 |
| chr17_+_34258411 | 0.09 |
ENSMUST00000087497.11
ENSMUST00000131134.9 ENSMUST00000235819.2 ENSMUST00000114255.9 ENSMUST00000114252.9 ENSMUST00000237989.2 |
Col11a2
|
collagen, type XI, alpha 2 |
| chr11_+_87628356 | 0.09 |
ENSMUST00000093955.12
|
Supt4a
|
SPT4A, DSIF elongation factor subunit |
| chr6_-_83808717 | 0.09 |
ENSMUST00000058383.9
|
Paip2b
|
poly(A) binding protein interacting protein 2B |
| chr8_+_53964721 | 0.09 |
ENSMUST00000211424.2
ENSMUST00000033920.6 |
Aga
|
aspartylglucosaminidase |
| chr9_-_57375269 | 0.09 |
ENSMUST00000215059.2
ENSMUST00000046587.8 ENSMUST00000214256.2 |
Scamp5
|
secretory carrier membrane protein 5 |
| chr5_-_35886605 | 0.09 |
ENSMUST00000070203.14
|
Sh3tc1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr10_-_126866658 | 0.09 |
ENSMUST00000120547.2
ENSMUST00000152054.8 |
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr4_-_57143437 | 0.08 |
ENSMUST00000095076.10
ENSMUST00000030142.4 |
Epb41l4b
|
erythrocyte membrane protein band 4.1 like 4b |
| chr4_-_152122891 | 0.07 |
ENSMUST00000030792.2
|
Tas1r1
|
taste receptor, type 1, member 1 |
| chr3_+_65435825 | 0.07 |
ENSMUST00000047906.10
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr2_+_130116357 | 0.07 |
ENSMUST00000136621.9
ENSMUST00000141872.2 |
Nop56
|
NOP56 ribonucleoprotein |
| chr3_+_89958940 | 0.07 |
ENSMUST00000159064.8
|
4933434E20Rik
|
RIKEN cDNA 4933434E20 gene |
| chr9_+_60701749 | 0.07 |
ENSMUST00000214354.2
ENSMUST00000050183.7 ENSMUST00000217656.2 |
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr7_-_4448631 | 0.07 |
ENSMUST00000008579.14
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr4_-_129452180 | 0.07 |
ENSMUST00000067240.11
|
Lck
|
lymphocyte protein tyrosine kinase |
| chr9_-_44253588 | 0.07 |
ENSMUST00000215091.2
|
Hmbs
|
hydroxymethylbilane synthase |
| chr15_+_100232810 | 0.07 |
ENSMUST00000075420.6
|
Mettl7a3
|
methyltransferase like 7A3 |
| chr9_-_83688294 | 0.06 |
ENSMUST00000034796.14
ENSMUST00000183614.2 |
Elovl4
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4 |
| chrX_-_56371953 | 0.06 |
ENSMUST00000176986.8
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr15_-_57755753 | 0.06 |
ENSMUST00000022993.7
|
Derl1
|
Der1-like domain family, member 1 |
| chr4_-_129452148 | 0.06 |
ENSMUST00000167288.8
ENSMUST00000134336.3 |
Lck
|
lymphocyte protein tyrosine kinase |
| chr16_+_18245134 | 0.06 |
ENSMUST00000205679.3
|
Txnrd2
|
thioredoxin reductase 2 |
| chr4_-_126362372 | 0.06 |
ENSMUST00000097888.10
ENSMUST00000239399.2 |
Ago1
|
argonaute RISC catalytic subunit 1 |
| chr7_+_130179063 | 0.05 |
ENSMUST00000207918.2
ENSMUST00000215492.2 ENSMUST00000084513.12 ENSMUST00000059145.14 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr7_-_84339045 | 0.05 |
ENSMUST00000209165.2
|
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr9_+_59564482 | 0.05 |
ENSMUST00000216620.2
ENSMUST00000217038.2 |
Pkm
|
pyruvate kinase, muscle |
| chr11_+_109541400 | 0.05 |
ENSMUST00000106676.8
|
Prkar1a
|
protein kinase, cAMP dependent regulatory, type I, alpha |
| chr8_-_4309257 | 0.05 |
ENSMUST00000053252.9
|
Ctxn1
|
cortexin 1 |
| chr11_-_82761954 | 0.05 |
ENSMUST00000108173.10
ENSMUST00000071152.14 |
Rffl
|
ring finger and FYVE like domain containing protein |
| chr8_-_78337297 | 0.05 |
ENSMUST00000141202.2
ENSMUST00000152168.8 |
Tmem184c
|
transmembrane protein 184C |
| chr2_+_29855572 | 0.05 |
ENSMUST00000113719.9
ENSMUST00000113717.8 ENSMUST00000113741.8 ENSMUST00000100225.9 ENSMUST00000095083.11 ENSMUST00000046257.14 |
Sptan1
|
spectrin alpha, non-erythrocytic 1 |
| chr7_-_126817639 | 0.04 |
ENSMUST00000152267.8
ENSMUST00000106314.8 |
Septin1
|
septin 1 |
| chr11_+_109541747 | 0.04 |
ENSMUST00000049527.7
|
Prkar1a
|
protein kinase, cAMP dependent regulatory, type I, alpha |
| chr13_+_55612050 | 0.04 |
ENSMUST00000046533.9
|
Prr7
|
proline rich 7 (synaptic) |
| chr8_-_106660470 | 0.04 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chr7_-_28931873 | 0.04 |
ENSMUST00000085818.6
|
Kcnk6
|
potassium inwardly-rectifying channel, subfamily K, member 6 |
| chr17_-_35978438 | 0.04 |
ENSMUST00000043674.15
|
Vars2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr4_-_135221926 | 0.04 |
ENSMUST00000102549.10
|
Nipal3
|
NIPA-like domain containing 3 |
| chr9_-_108444561 | 0.04 |
ENSMUST00000074208.6
|
Ndufaf3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr17_+_45874800 | 0.04 |
ENSMUST00000224905.2
ENSMUST00000226086.2 ENSMUST00000041353.7 |
Slc35b2
|
solute carrier family 35, member B2 |
| chr7_+_24310171 | 0.04 |
ENSMUST00000206422.2
|
Phldb3
|
pleckstrin homology like domain, family B, member 3 |
| chr8_-_117809188 | 0.04 |
ENSMUST00000109093.9
ENSMUST00000098375.6 |
Pkd1l2
|
polycystic kidney disease 1 like 2 |
| chr6_+_113366207 | 0.03 |
ENSMUST00000204026.3
|
Ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr1_+_171265103 | 0.03 |
ENSMUST00000043839.5
|
F11r
|
F11 receptor |
| chr17_+_31515962 | 0.03 |
ENSMUST00000237932.2
|
Slc37a1
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 1 |
| chr9_-_54569128 | 0.03 |
ENSMUST00000034822.12
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr19_-_11243530 | 0.03 |
ENSMUST00000169159.3
|
Ms4a1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr6_-_29164981 | 0.02 |
ENSMUST00000007993.16
|
Rbm28
|
RNA binding motif protein 28 |
| chr15_-_100497863 | 0.02 |
ENSMUST00000073837.13
|
Pou6f1
|
POU domain, class 6, transcription factor 1 |
| chr12_-_113945961 | 0.02 |
ENSMUST00000103466.2
|
Ighv11-1
|
immunoglobulin heavy variable 11-1 |
| chr4_+_118750282 | 0.02 |
ENSMUST00000094830.2
|
Olfr1330
|
olfactory receptor 1330 |
| chr13_-_104246084 | 0.02 |
ENSMUST00000224945.2
ENSMUST00000109315.5 |
Nln
|
neurolysin (metallopeptidase M3 family) |
| chr11_+_99748741 | 0.02 |
ENSMUST00000107434.2
|
Gm11568
|
predicted gene 11568 |
| chr16_+_31482745 | 0.02 |
ENSMUST00000100001.10
ENSMUST00000064477.14 |
Dlg1
|
discs large MAGUK scaffold protein 1 |
| chr7_-_4448180 | 0.02 |
ENSMUST00000138798.2
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr9_-_14815228 | 0.02 |
ENSMUST00000034409.14
ENSMUST00000117620.8 |
Izumo1r
|
IZUMO1 receptor, JUNO |
| chr7_+_30121776 | 0.02 |
ENSMUST00000108175.8
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr5_-_23880939 | 0.02 |
ENSMUST00000196388.5
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr11_-_109188892 | 0.01 |
ENSMUST00000106706.8
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr12_+_102915102 | 0.01 |
ENSMUST00000101099.12
|
Unc79
|
unc-79 homolog |
| chr5_+_8943943 | 0.01 |
ENSMUST00000196067.2
|
Abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr11_-_109188947 | 0.01 |
ENSMUST00000020920.10
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr11_+_102993865 | 0.01 |
ENSMUST00000152971.2
|
Acbd4
|
acyl-Coenzyme A binding domain containing 4 |
| chr11_-_99549786 | 0.01 |
ENSMUST00000092695.5
|
Gm11563
|
predicted gene 11563 |
| chr7_-_142628298 | 0.01 |
ENSMUST00000148715.8
|
Trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr8_+_55024446 | 0.01 |
ENSMUST00000239166.2
ENSMUST00000239106.2 ENSMUST00000239152.2 |
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr9_+_118307918 | 0.01 |
ENSMUST00000150633.2
|
Eomes
|
eomesodermin |
| chr6_+_83112793 | 0.01 |
ENSMUST00000065512.11
|
Rtkn
|
rhotekin |
| chr11_-_99556781 | 0.01 |
ENSMUST00000100476.3
|
Krtap4-6
|
keratin associated protein 4-6 |
| chr19_+_38995463 | 0.01 |
ENSMUST00000025966.5
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55 |
| chr11_+_99839246 | 0.01 |
ENSMUST00000105052.2
|
Krtap9-5
|
keratin associated protein 9-5 |
| chr17_+_21534225 | 0.00 |
ENSMUST00000077301.6
|
Vmn1r237
|
vomeronasal 1 receptor 237 |
| chr11_-_99684173 | 0.00 |
ENSMUST00000105058.2
|
Gm11596
|
predicted gene 11596 |
| chr8_+_106032205 | 0.00 |
ENSMUST00000109375.4
ENSMUST00000212033.2 |
Elmo3
|
engulfment and cell motility 3 |
| chr6_+_40941688 | 0.00 |
ENSMUST00000076638.7
|
1810009J06Rik
|
RIKEN cDNA 1810009J06 gene |
| chr11_-_99689758 | 0.00 |
ENSMUST00000105057.2
|
Gm11569
|
predicted gene 11569 |
| chr9_-_14815050 | 0.00 |
ENSMUST00000148155.2
ENSMUST00000121116.8 |
Izumo1r
|
IZUMO1 receptor, JUNO |
| chr7_-_24919247 | 0.00 |
ENSMUST00000058702.7
|
Dedd2
|
death effector domain-containing DNA binding protein 2 |
| chr6_-_40976413 | 0.00 |
ENSMUST00000166306.3
|
Gm2663
|
predicted gene 2663 |
| chr16_-_17394080 | 0.00 |
ENSMUST00000231552.2
|
Slc7a4
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 4 |
| chr9_-_114673158 | 0.00 |
ENSMUST00000047013.4
|
Cmtm8
|
CKLF-like MARVEL transmembrane domain containing 8 |
| chr9_-_54568950 | 0.00 |
ENSMUST00000128624.2
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr15_-_38300937 | 0.00 |
ENSMUST00000227920.2
ENSMUST00000074043.7 ENSMUST00000228416.2 |
Klf10
|
Kruppel-like factor 10 |
| chr9_-_14815163 | 0.00 |
ENSMUST00000069408.10
|
Izumo1r
|
IZUMO1 receptor, JUNO |
| chr2_-_79959802 | 0.00 |
ENSMUST00000102653.8
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chrX_-_52203405 | 0.00 |
ENSMUST00000114843.9
|
Plac1
|
placental specific protein 1 |
| chr13_+_104246259 | 0.00 |
ENSMUST00000160322.8
ENSMUST00000159574.2 |
Sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr8_-_112120442 | 0.00 |
ENSMUST00000038475.9
|
Fa2h
|
fatty acid 2-hydroxylase |
| chr11_-_99742434 | 0.00 |
ENSMUST00000107437.2
|
Krtap4-16
|
keratin associated protein 4-16 |
| chr11_+_69826719 | 0.00 |
ENSMUST00000149194.8
|
Ybx2
|
Y box protein 2 |
| chr5_+_24569802 | 0.00 |
ENSMUST00000115090.6
ENSMUST00000030834.7 |
Nos3
|
nitric oxide synthase 3, endothelial cell |
| chr2_+_139520098 | 0.00 |
ENSMUST00000184404.8
ENSMUST00000099307.4 |
Ism1
|
isthmin 1, angiogenesis inhibitor |
Network of associatons between targets according to the STRING database.
First level regulatory network of Twist1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.3 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.1 | 0.2 | GO:0019405 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.1 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.0 | 0.2 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0032831 | positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.0 | 0.2 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0009644 | response to high light intensity(GO:0009644) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.1 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.1 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.0 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |