Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
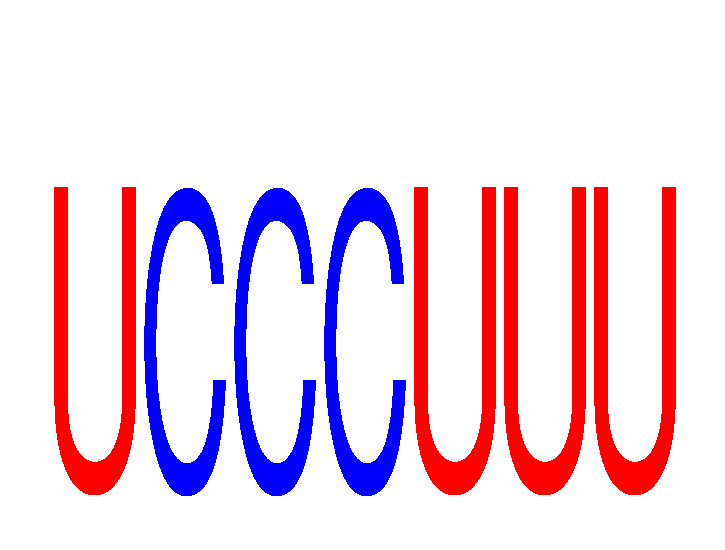
Results for UCCCUUU
Z-value: 0.40
miRNA associated with seed UCCCUUU
| Name | miRBASE accession |
|---|---|
|
mmu-miR-204-5p
|
MIMAT0000237 |
|
mmu-miR-211-5p
|
MIMAT0000668 |
|
mmu-miR-7670-3p
|
MIMAT0029847 |
Activity profile of UCCCUUU motif
Sorted Z-values of UCCCUUU motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_9550907 | 0.14 |
ENSMUST00000100070.5
|
Samd5
|
sterile alpha motif domain containing 5 |
| chr14_+_121272950 | 0.12 |
ENSMUST00000026635.8
|
Farp1
|
FERM, RhoGEF (Arhgef) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr15_-_90563510 | 0.09 |
ENSMUST00000014777.9
ENSMUST00000064391.12 |
Cpne8
|
copine VIII |
| chr1_-_79649683 | 0.08 |
ENSMUST00000162342.8
|
Ap1s3
|
adaptor-related protein complex AP-1, sigma 3 |
| chr17_-_12988492 | 0.08 |
ENSMUST00000024599.14
|
Igf2r
|
insulin-like growth factor 2 receptor |
| chr10_-_84276454 | 0.08 |
ENSMUST00000020220.15
|
Nuak1
|
NUAK family, SNF1-like kinase, 1 |
| chr4_-_3938352 | 0.07 |
ENSMUST00000003369.10
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chr14_-_25769457 | 0.07 |
ENSMUST00000069180.8
|
Zcchc24
|
zinc finger, CCHC domain containing 24 |
| chr2_+_84670956 | 0.07 |
ENSMUST00000111625.2
|
Slc43a1
|
solute carrier family 43, member 1 |
| chr16_+_8647959 | 0.07 |
ENSMUST00000023150.7
|
1810013L24Rik
|
RIKEN cDNA 1810013L24 gene |
| chr8_+_109441276 | 0.06 |
ENSMUST00000043896.10
|
Zfhx3
|
zinc finger homeobox 3 |
| chr18_-_16942289 | 0.06 |
ENSMUST00000025166.14
|
Cdh2
|
cadherin 2 |
| chr14_+_49303952 | 0.06 |
ENSMUST00000037473.6
ENSMUST00000228238.2 |
Ap5m1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr17_+_81251997 | 0.06 |
ENSMUST00000025092.5
|
Tmem178
|
transmembrane protein 178 |
| chr3_-_108819003 | 0.06 |
ENSMUST00000029480.9
|
Prpf38b
|
PRP38 pre-mRNA processing factor 38 (yeast) domain containing B |
| chr15_+_25622611 | 0.06 |
ENSMUST00000110457.8
ENSMUST00000137601.8 |
Myo10
|
myosin X |
| chr5_+_123532819 | 0.05 |
ENSMUST00000111596.8
ENSMUST00000068237.12 |
Mlxip
|
MLX interacting protein |
| chr5_-_51711237 | 0.05 |
ENSMUST00000132734.8
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr1_-_106641940 | 0.05 |
ENSMUST00000112751.2
|
Bcl2
|
B cell leukemia/lymphoma 2 |
| chr13_+_41013230 | 0.05 |
ENSMUST00000110191.10
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr14_-_39194782 | 0.04 |
ENSMUST00000168810.9
ENSMUST00000173780.2 ENSMUST00000166968.9 |
Nrg3
|
neuregulin 3 |
| chr6_+_47854138 | 0.04 |
ENSMUST00000061890.8
|
Zfp282
|
zinc finger protein 282 |
| chr6_-_39354570 | 0.04 |
ENSMUST00000200771.4
ENSMUST00000202204.4 ENSMUST00000202952.2 ENSMUST00000202400.4 ENSMUST00000200969.4 ENSMUST00000202749.4 ENSMUST00000090243.8 |
Slc37a3
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 3 |
| chr2_+_3714492 | 0.04 |
ENSMUST00000027965.11
|
Fam107b
|
family with sequence similarity 107, member B |
| chr7_+_141142075 | 0.04 |
ENSMUST00000003038.12
|
Ap2a2
|
adaptor-related protein complex 2, alpha 2 subunit |
| chr9_-_60418286 | 0.04 |
ENSMUST00000098660.10
|
Thsd4
|
thrombospondin, type I, domain containing 4 |
| chr5_-_62923463 | 0.04 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr2_-_5900130 | 0.04 |
ENSMUST00000026926.5
ENSMUST00000193792.6 ENSMUST00000102981.10 |
Sec61a2
|
Sec61, alpha subunit 2 (S. cerevisiae) |
| chr15_-_98505508 | 0.04 |
ENSMUST00000096224.6
|
Adcy6
|
adenylate cyclase 6 |
| chr10_+_67815395 | 0.04 |
ENSMUST00000118160.8
|
Rtkn2
|
rhotekin 2 |
| chr5_-_114046746 | 0.04 |
ENSMUST00000004646.13
|
Coro1c
|
coronin, actin binding protein 1C |
| chr7_+_4122523 | 0.04 |
ENSMUST00000119661.8
ENSMUST00000129423.8 |
Ttyh1
|
tweety family member 1 |
| chr15_-_78004211 | 0.03 |
ENSMUST00000019290.3
|
Cacng2
|
calcium channel, voltage-dependent, gamma subunit 2 |
| chr8_+_26609384 | 0.03 |
ENSMUST00000014022.15
ENSMUST00000209300.2 ENSMUST00000153528.8 ENSMUST00000209707.2 |
Rnf170
|
ring finger protein 170 |
| chr1_+_127701901 | 0.03 |
ENSMUST00000112570.2
ENSMUST00000027587.15 |
Ccnt2
|
cyclin T2 |
| chr8_-_26609153 | 0.03 |
ENSMUST00000037182.14
|
Hook3
|
hook microtubule tethering protein 3 |
| chr4_+_137004793 | 0.03 |
ENSMUST00000045747.5
|
Wnt4
|
wingless-type MMTV integration site family, member 4 |
| chr9_+_103879745 | 0.03 |
ENSMUST00000035167.15
ENSMUST00000194774.6 |
Nphp3
|
nephronophthisis 3 (adolescent) |
| chr1_-_34882131 | 0.03 |
ENSMUST00000167518.8
ENSMUST00000047534.12 |
Fam168b
|
family with sequence similarity 168, member B |
| chr9_-_106764627 | 0.03 |
ENSMUST00000055843.9
|
Rbm15b
|
RNA binding motif protein 15B |
| chr9_-_21963306 | 0.03 |
ENSMUST00000003501.9
ENSMUST00000215901.2 |
Elavl3
|
ELAV like RNA binding protein 3 |
| chr13_+_31990604 | 0.03 |
ENSMUST00000062292.5
|
Foxc1
|
forkhead box C1 |
| chr15_-_98796373 | 0.03 |
ENSMUST00000229775.2
ENSMUST00000023737.6 |
Dhh
|
desert hedgehog |
| chr16_+_42727926 | 0.03 |
ENSMUST00000151244.8
ENSMUST00000114694.9 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr6_-_31540913 | 0.03 |
ENSMUST00000026698.8
|
Podxl
|
podocalyxin-like |
| chr3_+_129326004 | 0.03 |
ENSMUST00000199910.5
ENSMUST00000197070.5 ENSMUST00000071402.7 |
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr1_-_43137768 | 0.03 |
ENSMUST00000190427.7
ENSMUST00000095014.8 |
Tgfbrap1
|
transforming growth factor, beta receptor associated protein 1 |
| chr6_+_100681670 | 0.03 |
ENSMUST00000032157.9
|
Gxylt2
|
glucoside xylosyltransferase 2 |
| chr1_+_160733942 | 0.03 |
ENSMUST00000161609.8
|
Rc3h1
|
RING CCCH (C3H) domains 1 |
| chr3_-_95125002 | 0.02 |
ENSMUST00000107209.8
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr8_-_124709859 | 0.02 |
ENSMUST00000075578.7
|
Abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr14_-_21102487 | 0.02 |
ENSMUST00000154460.8
ENSMUST00000130291.8 |
Ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr9_+_56982622 | 0.02 |
ENSMUST00000167715.8
|
Sin3a
|
transcriptional regulator, SIN3A (yeast) |
| chr12_-_84265609 | 0.02 |
ENSMUST00000046266.13
ENSMUST00000220974.2 |
Mideas
|
mitotic deacetylase associated SANT domain protein |
| chr3_+_89325750 | 0.02 |
ENSMUST00000039110.12
ENSMUST00000125036.8 ENSMUST00000191485.7 ENSMUST00000154791.8 |
Shc1
|
src homology 2 domain-containing transforming protein C1 |
| chr13_+_9143995 | 0.02 |
ENSMUST00000091829.4
|
Larp4b
|
La ribonucleoprotein domain family, member 4B |
| chr7_+_99184645 | 0.02 |
ENSMUST00000098266.9
ENSMUST00000179755.8 |
Arrb1
|
arrestin, beta 1 |
| chr2_-_130681978 | 0.02 |
ENSMUST00000044766.15
ENSMUST00000138990.2 ENSMUST00000120316.8 ENSMUST00000110243.8 |
4930402H24Rik
|
RIKEN cDNA 4930402H24 gene |
| chr3_-_97134680 | 0.02 |
ENSMUST00000046521.14
|
Bcl9
|
B cell CLL/lymphoma 9 |
| chr10_-_75353157 | 0.02 |
ENSMUST00000039796.14
|
Gucd1
|
guanylyl cyclase domain containing 1 |
| chrX_-_63320543 | 0.02 |
ENSMUST00000114679.2
ENSMUST00000069926.14 |
Slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr11_+_97253221 | 0.02 |
ENSMUST00000238729.2
ENSMUST00000045540.4 |
Socs7
|
suppressor of cytokine signaling 7 |
| chr15_+_80595486 | 0.02 |
ENSMUST00000067689.9
|
Tnrc6b
|
trinucleotide repeat containing 6b |
| chr1_-_84818223 | 0.02 |
ENSMUST00000186465.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr5_+_110324506 | 0.02 |
ENSMUST00000112512.8
|
Golga3
|
golgi autoantigen, golgin subfamily a, 3 |
| chr2_+_173501553 | 0.02 |
ENSMUST00000029024.10
ENSMUST00000109110.4 |
Rab22a
|
RAB22A, member RAS oncogene family |
| chr7_+_127344942 | 0.02 |
ENSMUST00000189562.7
ENSMUST00000186116.7 |
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr10_+_22520910 | 0.02 |
ENSMUST00000042261.5
|
Slc2a12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr4_-_142939260 | 0.02 |
ENSMUST00000105778.8
ENSMUST00000134791.2 |
Prdm2
|
PR domain containing 2, with ZNF domain |
| chr5_+_108609087 | 0.02 |
ENSMUST00000112597.8
ENSMUST00000046975.12 |
Pcgf3
|
polycomb group ring finger 3 |
| chr12_-_107969853 | 0.02 |
ENSMUST00000066060.11
|
Bcl11b
|
B cell leukemia/lymphoma 11B |
| chr3_-_89820451 | 0.02 |
ENSMUST00000029559.7
ENSMUST00000197679.5 |
Il6ra
|
interleukin 6 receptor, alpha |
| chr13_-_76091931 | 0.02 |
ENSMUST00000022078.12
ENSMUST00000109606.3 |
Rhobtb3
|
Rho-related BTB domain containing 3 |
| chr2_+_15060051 | 0.02 |
ENSMUST00000069870.11
ENSMUST00000239125.2 ENSMUST00000193836.3 |
Arl5b
|
ADP-ribosylation factor-like 5B |
| chr11_-_18968979 | 0.02 |
ENSMUST00000144988.8
|
Meis1
|
Meis homeobox 1 |
| chr2_+_116951855 | 0.01 |
ENSMUST00000028829.13
|
Spred1
|
sprouty protein with EVH-1 domain 1, related sequence |
| chr16_-_25924527 | 0.01 |
ENSMUST00000039990.6
|
P3h2
|
prolyl 3-hydroxylase 2 |
| chr2_-_164753480 | 0.01 |
ENSMUST00000041361.14
|
Zfp335
|
zinc finger protein 335 |
| chr12_-_34578842 | 0.01 |
ENSMUST00000110819.4
|
Hdac9
|
histone deacetylase 9 |
| chr12_+_76417040 | 0.01 |
ENSMUST00000042779.4
|
Zbtb1
|
zinc finger and BTB domain containing 1 |
| chr4_+_150366028 | 0.01 |
ENSMUST00000105682.9
|
Rere
|
arginine glutamic acid dipeptide (RE) repeats |
| chr2_-_52314837 | 0.01 |
ENSMUST00000036541.8
|
Arl5a
|
ADP-ribosylation factor-like 5A |
| chr5_-_99400698 | 0.01 |
ENSMUST00000031276.15
ENSMUST00000168092.8 |
Rasgef1b
|
RasGEF domain family, member 1B |
| chr6_-_57512355 | 0.01 |
ENSMUST00000042766.6
|
Ppm1k
|
protein phosphatase 1K (PP2C domain containing) |
| chr13_+_74157538 | 0.01 |
ENSMUST00000022057.9
|
Tppp
|
tubulin polymerization promoting protein |
| chr4_+_57637817 | 0.01 |
ENSMUST00000150412.4
|
Pakap
|
paralemmin A kinase anchor protein |
| chr9_+_62585108 | 0.01 |
ENSMUST00000034774.9
|
Itga11
|
integrin alpha 11 |
| chr8_+_106786190 | 0.01 |
ENSMUST00000109308.3
|
Nfatc3
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
| chr19_-_41969558 | 0.01 |
ENSMUST00000026168.9
ENSMUST00000171561.8 |
Mms19
|
MMS19 cytosolic iron-sulfur assembly component |
| chr7_-_105049197 | 0.01 |
ENSMUST00000048079.14
ENSMUST00000118726.8 ENSMUST00000074686.10 ENSMUST00000122327.7 ENSMUST00000179474.9 |
Fam160a2
|
family with sequence similarity 160, member A2 |
| chr4_+_57845240 | 0.01 |
ENSMUST00000102903.8
ENSMUST00000107598.9 |
Pakap
|
paralemmin A kinase anchor protein |
| chr12_-_46865709 | 0.01 |
ENSMUST00000021438.8
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr1_-_190643922 | 0.01 |
ENSMUST00000159066.2
ENSMUST00000061611.15 |
Rps6kc1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr2_-_69416365 | 0.01 |
ENSMUST00000100051.9
ENSMUST00000092551.5 ENSMUST00000080953.12 |
Lrp2
|
low density lipoprotein receptor-related protein 2 |
| chr14_-_61794330 | 0.01 |
ENSMUST00000022497.15
|
Spryd7
|
SPRY domain containing 7 |
| chr17_-_45047521 | 0.01 |
ENSMUST00000113572.9
|
Runx2
|
runt related transcription factor 2 |
| chr11_+_72580823 | 0.01 |
ENSMUST00000155998.2
|
Ankfy1
|
ankyrin repeat and FYVE domain containing 1 |
| chr6_-_92191878 | 0.01 |
ENSMUST00000014694.11
|
Rbsn
|
rabenosyn, RAB effector |
| chr11_-_69451012 | 0.01 |
ENSMUST00000004036.6
|
Efnb3
|
ephrin B3 |
| chr10_+_127928622 | 0.01 |
ENSMUST00000219072.2
ENSMUST00000045621.9 ENSMUST00000170054.9 |
Baz2a
|
bromodomain adjacent to zinc finger domain, 2A |
| chr1_-_57011595 | 0.01 |
ENSMUST00000042857.14
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr15_+_6737853 | 0.01 |
ENSMUST00000061656.8
|
Rictor
|
RPTOR independent companion of MTOR, complex 2 |
| chr3_-_138195133 | 0.01 |
ENSMUST00000197531.2
ENSMUST00000029804.13 |
Metap1
|
methionyl aminopeptidase 1 |
| chr4_+_117109204 | 0.01 |
ENSMUST00000125943.8
ENSMUST00000106434.8 |
Tmem53
|
transmembrane protein 53 |
| chr7_-_128063354 | 0.01 |
ENSMUST00000106226.9
ENSMUST00000106228.8 |
Tial1
|
Tia1 cytotoxic granule-associated RNA binding protein-like 1 |
| chr13_-_29137673 | 0.01 |
ENSMUST00000067230.6
|
Sox4
|
SRY (sex determining region Y)-box 4 |
| chr10_+_4561974 | 0.01 |
ENSMUST00000105590.8
ENSMUST00000067086.14 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr13_+_38010203 | 0.01 |
ENSMUST00000128570.9
|
Rreb1
|
ras responsive element binding protein 1 |
| chr10_+_126914755 | 0.01 |
ENSMUST00000039259.7
ENSMUST00000217941.2 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr16_+_17269845 | 0.01 |
ENSMUST00000006293.5
ENSMUST00000231228.2 |
Crkl
|
v-crk avian sarcoma virus CT10 oncogene homolog-like |
| chrX_+_162691978 | 0.01 |
ENSMUST00000069041.15
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr5_-_135963408 | 0.01 |
ENSMUST00000198270.2
ENSMUST00000055808.6 |
Ywhag
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide |
| chr11_+_80099843 | 0.01 |
ENSMUST00000077451.14
ENSMUST00000055056.16 |
Rhot1
|
ras homolog family member T1 |
| chr2_-_156833932 | 0.01 |
ENSMUST00000109558.2
ENSMUST00000069600.13 ENSMUST00000072298.13 |
Ndrg3
|
N-myc downstream regulated gene 3 |
| chr3_-_94566107 | 0.01 |
ENSMUST00000196655.5
ENSMUST00000200407.2 ENSMUST00000006123.11 ENSMUST00000196733.5 |
Tuft1
|
tuftelin 1 |
| chr2_-_57004933 | 0.01 |
ENSMUST00000028166.9
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr10_-_122912272 | 0.01 |
ENSMUST00000219203.2
ENSMUST00000073792.11 |
Mon2
|
MON2 homolog, regulator of endosome to Golgi trafficking |
| chr4_-_58553311 | 0.01 |
ENSMUST00000107571.8
ENSMUST00000055018.11 |
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr9_+_95836797 | 0.01 |
ENSMUST00000034981.14
ENSMUST00000185633.7 |
Xrn1
|
5'-3' exoribonuclease 1 |
| chr2_+_31462780 | 0.01 |
ENSMUST00000137889.7
ENSMUST00000194386.6 ENSMUST00000055244.13 |
Fubp3
|
far upstream element (FUSE) binding protein 3 |
| chrX_-_55643429 | 0.01 |
ENSMUST00000059899.3
|
Mmgt1
|
membrane magnesium transporter 1 |
| chr1_-_21149392 | 0.01 |
ENSMUST00000037998.6
|
Tram2
|
translocating chain-associating membrane protein 2 |
| chr19_+_5524701 | 0.01 |
ENSMUST00000165485.8
ENSMUST00000166253.8 ENSMUST00000167371.8 ENSMUST00000167855.8 ENSMUST00000070118.14 |
Efemp2
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 2 |
| chr18_-_39623698 | 0.01 |
ENSMUST00000115567.8
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr1_-_163141230 | 0.01 |
ENSMUST00000174397.3
ENSMUST00000075805.13 |
Prrx1
|
paired related homeobox 1 |
| chr2_-_33321306 | 0.01 |
ENSMUST00000113158.8
|
Zbtb34
|
zinc finger and BTB domain containing 34 |
| chrX_+_100892981 | 0.01 |
ENSMUST00000124279.6
ENSMUST00000101339.8 |
Nhsl2
|
NHS-like 2 |
| chr10_-_110845860 | 0.01 |
ENSMUST00000041723.15
|
Zdhhc17
|
zinc finger, DHHC domain containing 17 |
| chr18_-_15196612 | 0.01 |
ENSMUST00000168989.9
|
Kctd1
|
potassium channel tetramerisation domain containing 1 |
| chrX_-_143471176 | 0.01 |
ENSMUST00000040184.4
|
Trpc5
|
transient receptor potential cation channel, subfamily C, member 5 |
| chr5_+_124577952 | 0.00 |
ENSMUST00000059580.11
|
Kmt5a
|
lysine methyltransferase 5A |
| chr15_-_82796308 | 0.00 |
ENSMUST00000109510.10
ENSMUST00000048966.7 |
Tcf20
|
transcription factor 20 |
| chr1_-_170695328 | 0.00 |
ENSMUST00000027974.7
|
Atf6
|
activating transcription factor 6 |
| chr7_-_131012202 | 0.00 |
ENSMUST00000207243.2
ENSMUST00000128432.3 ENSMUST00000121033.8 ENSMUST00000046306.15 |
Ikzf5
|
IKAROS family zinc finger 5 |
| chr19_-_18978463 | 0.00 |
ENSMUST00000040153.15
ENSMUST00000112828.8 |
Rorb
|
RAR-related orphan receptor beta |
| chr16_+_56942050 | 0.00 |
ENSMUST00000166897.3
|
Tomm70a
|
translocase of outer mitochondrial membrane 70A |
| chr19_+_41471067 | 0.00 |
ENSMUST00000067795.13
|
Lcor
|
ligand dependent nuclear receptor corepressor |
| chr18_+_69478790 | 0.00 |
ENSMUST00000202116.4
ENSMUST00000114982.8 ENSMUST00000078486.13 ENSMUST00000202772.4 ENSMUST00000201288.4 |
Tcf4
|
transcription factor 4 |
| chr7_+_132460954 | 0.00 |
ENSMUST00000084497.12
ENSMUST00000106161.8 |
Abraxas2
|
BRISC complex subunit |
| chr10_-_116984386 | 0.00 |
ENSMUST00000020381.5
|
Frs2
|
fibroblast growth factor receptor substrate 2 |
| chr17_-_80787398 | 0.00 |
ENSMUST00000068714.7
|
Sos1
|
SOS Ras/Rac guanine nucleotide exchange factor 1 |
| chr4_-_155306992 | 0.00 |
ENSMUST00000084103.10
ENSMUST00000030917.6 |
Ski
|
ski sarcoma viral oncogene homolog (avian) |
| chr2_-_34645241 | 0.00 |
ENSMUST00000102800.9
|
Gapvd1
|
GTPase activating protein and VPS9 domains 1 |
| chr2_-_33777874 | 0.00 |
ENSMUST00000041555.10
|
Mvb12b
|
multivesicular body subunit 12B |
| chr10_+_128641520 | 0.00 |
ENSMUST00000219508.2
ENSMUST00000026410.2 |
Dnajc14
|
DnaJ heat shock protein family (Hsp40) member C14 |
| chr6_-_92920466 | 0.00 |
ENSMUST00000113438.8
|
Adamts9
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 9 |
| chr2_-_37537224 | 0.00 |
ENSMUST00000028279.10
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr11_+_105975403 | 0.00 |
ENSMUST00000002044.10
|
Map3k3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr8_-_122379631 | 0.00 |
ENSMUST00000046386.5
|
Zcchc14
|
zinc finger, CCHC domain containing 14 |
| chr4_+_57434247 | 0.00 |
ENSMUST00000102905.8
|
Pakap
|
paralemmin A kinase anchor protein |
| chr19_-_7218512 | 0.00 |
ENSMUST00000025675.11
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr11_+_54329014 | 0.00 |
ENSMUST00000046835.14
|
Fnip1
|
folliculin interacting protein 1 |
| chr1_+_59952131 | 0.00 |
ENSMUST00000036540.12
|
Fam117b
|
family with sequence similarity 117, member B |
| chr14_+_55120875 | 0.00 |
ENSMUST00000134077.2
ENSMUST00000172844.8 ENSMUST00000133397.4 ENSMUST00000227108.2 |
Gm20521
Bcl2l2
|
predicted gene 20521 BCL2-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of UCCCUUU
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1903210 | glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.0 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.0 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.0 | GO:0072034 | positive regulation of dermatome development(GO:0061184) renal vesicle induction(GO:0072034) negative regulation of male gonad development(GO:2000019) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990844 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |