Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
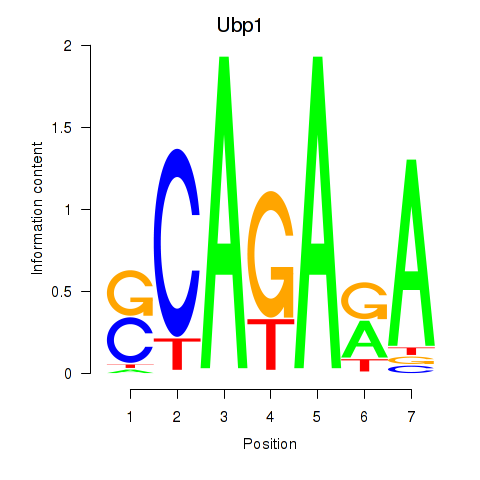
Results for Ubp1
Z-value: 1.04
Transcription factors associated with Ubp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ubp1
|
ENSMUSG00000009741.16 | upstream binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ubp1 | mm39_v1_chr9_+_113760376_113760722 | -0.57 | 3.1e-01 | Click! |
Activity profile of Ubp1 motif
Sorted Z-values of Ubp1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_106020545 | 1.02 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chr18_-_43610829 | 0.82 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr10_-_128384971 | 0.81 |
ENSMUST00000176906.2
|
Rpl41
|
ribosomal protein L41 |
| chr17_-_36440317 | 0.59 |
ENSMUST00000046131.16
ENSMUST00000173322.8 ENSMUST00000172968.2 |
Gm7030
|
predicted gene 7030 |
| chr8_-_79539838 | 0.52 |
ENSMUST00000146824.2
|
Lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr6_+_41523664 | 0.49 |
ENSMUST00000103299.3
|
Trbc2
|
T cell receptor beta, constant 2 |
| chr14_-_56339915 | 0.43 |
ENSMUST00000015583.2
|
Ctsg
|
cathepsin G |
| chr2_-_26800581 | 0.40 |
ENSMUST00000015920.12
ENSMUST00000139815.2 ENSMUST00000102899.10 |
Med22
|
mediator complex subunit 22 |
| chr1_+_172327812 | 0.39 |
ENSMUST00000192460.2
|
Tagln2
|
transgelin 2 |
| chr16_-_35311243 | 0.39 |
ENSMUST00000023550.9
|
Pdia5
|
protein disulfide isomerase associated 5 |
| chr2_+_80145805 | 0.37 |
ENSMUST00000028392.8
|
Dnajc10
|
DnaJ heat shock protein family (Hsp40) member C10 |
| chr6_+_116528102 | 0.37 |
ENSMUST00000122096.3
|
Eif4a3l2
|
eukaryotic translation initiation factor 4A3 like 2 |
| chr10_+_75768964 | 0.37 |
ENSMUST00000219839.2
|
Chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr2_-_65955338 | 0.37 |
ENSMUST00000028378.4
|
Galnt3
|
polypeptide N-acetylgalactosaminyltransferase 3 |
| chr10_-_85793639 | 0.36 |
ENSMUST00000001834.4
|
Rtcb
|
RNA 2',3'-cyclic phosphate and 5'-OH ligase |
| chr6_-_52217821 | 0.36 |
ENSMUST00000121043.2
|
Hoxa10
|
homeobox A10 |
| chrX_+_93679671 | 0.35 |
ENSMUST00000096368.4
|
Gspt2
|
G1 to S phase transition 2 |
| chr10_+_19497740 | 0.35 |
ENSMUST00000036564.8
|
Il22ra2
|
interleukin 22 receptor, alpha 2 |
| chr2_-_164198427 | 0.35 |
ENSMUST00000109367.10
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr19_-_4092218 | 0.34 |
ENSMUST00000237999.2
ENSMUST00000042700.12 |
Gstp2
|
glutathione S-transferase, pi 2 |
| chr18_+_78392969 | 0.33 |
ENSMUST00000164064.2
|
Gm6133
|
predicted gene 6133 |
| chr10_-_128384994 | 0.33 |
ENSMUST00000177163.8
ENSMUST00000176683.8 ENSMUST00000176010.8 |
Rpl41
|
ribosomal protein L41 |
| chr17_-_27816151 | 0.33 |
ENSMUST00000231742.2
|
Nudt3
|
nudix (nucleotide diphosphate linked moiety X)-type motif 3 |
| chr15_-_79976016 | 0.32 |
ENSMUST00000185306.3
|
Rpl3
|
ribosomal protein L3 |
| chr4_-_156340276 | 0.32 |
ENSMUST00000220228.2
ENSMUST00000218788.2 ENSMUST00000179919.3 |
Samd11
|
sterile alpha motif domain containing 11 |
| chr11_+_62770275 | 0.32 |
ENSMUST00000014321.5
|
Tvp23b
|
trans-golgi network vesicle protein 23B |
| chr17_+_29309942 | 0.31 |
ENSMUST00000119901.9
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr2_-_112089627 | 0.31 |
ENSMUST00000043970.2
|
Nutm1
|
NUT midline carcinoma, family member 1 |
| chr16_+_48692976 | 0.31 |
ENSMUST00000065666.6
|
Retnlg
|
resistin like gamma |
| chr17_+_26882171 | 0.30 |
ENSMUST00000236346.2
|
Atp6v0e
|
ATPase, H+ transporting, lysosomal V0 subunit E |
| chr1_+_166206666 | 0.30 |
ENSMUST00000027846.8
|
Tada1
|
transcriptional adaptor 1 |
| chr3_+_5815863 | 0.30 |
ENSMUST00000192045.2
|
Gm8797
|
predicted pseudogene 8797 |
| chr4_+_140427799 | 0.30 |
ENSMUST00000071169.9
|
Rcc2
|
regulator of chromosome condensation 2 |
| chr11_+_68961599 | 0.29 |
ENSMUST00000075980.12
ENSMUST00000094081.5 |
Tmem107
|
transmembrane protein 107 |
| chr4_+_119052693 | 0.29 |
ENSMUST00000097908.4
|
Svbp
|
small vasohibin binding protein |
| chr1_+_79753735 | 0.29 |
ENSMUST00000027464.9
|
Mrpl44
|
mitochondrial ribosomal protein L44 |
| chr7_+_46490899 | 0.29 |
ENSMUST00000147535.8
|
Ldha
|
lactate dehydrogenase A |
| chr16_+_20317544 | 0.29 |
ENSMUST00000003320.14
|
Eif2b5
|
eukaryotic translation initiation factor 2B, subunit 5 epsilon |
| chr15_-_79967543 | 0.29 |
ENSMUST00000081650.15
|
Rpl3
|
ribosomal protein L3 |
| chr13_-_55676334 | 0.28 |
ENSMUST00000047877.5
|
Dok3
|
docking protein 3 |
| chr18_-_36859732 | 0.28 |
ENSMUST00000061829.8
|
Cd14
|
CD14 antigen |
| chr19_+_44282113 | 0.28 |
ENSMUST00000026221.7
|
Scd2
|
stearoyl-Coenzyme A desaturase 2 |
| chr2_+_103799873 | 0.28 |
ENSMUST00000123437.8
|
Lmo2
|
LIM domain only 2 |
| chr6_-_129252396 | 0.28 |
ENSMUST00000032259.6
|
Cd69
|
CD69 antigen |
| chr6_+_85429023 | 0.28 |
ENSMUST00000204592.3
|
Cct7
|
chaperonin containing Tcp1, subunit 7 (eta) |
| chr7_-_126303887 | 0.27 |
ENSMUST00000131415.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr3_-_57559088 | 0.27 |
ENSMUST00000160959.8
|
Commd2
|
COMM domain containing 2 |
| chr12_-_31684588 | 0.27 |
ENSMUST00000020979.9
ENSMUST00000177962.9 |
Bcap29
|
B cell receptor associated protein 29 |
| chr3_+_104545974 | 0.27 |
ENSMUST00000046212.2
|
Slc16a1
|
solute carrier family 16 (monocarboxylic acid transporters), member 1 |
| chr14_-_76248274 | 0.27 |
ENSMUST00000088922.5
|
Gtf2f2
|
general transcription factor IIF, polypeptide 2 |
| chr3_-_105940130 | 0.27 |
ENSMUST00000200146.2
|
Chil5
|
chitinase-like 5 |
| chr11_-_50093781 | 0.27 |
ENSMUST00000136936.2
|
Sqstm1
|
sequestosome 1 |
| chr1_-_44157916 | 0.27 |
ENSMUST00000027213.14
ENSMUST00000065767.9 |
Poglut2
|
protein O-glucosyltransferase 2 |
| chr6_+_134988572 | 0.26 |
ENSMUST00000032326.11
ENSMUST00000130851.8 ENSMUST00000205244.3 ENSMUST00000205055.3 ENSMUST00000204646.3 ENSMUST00000154558.3 |
Ddx47
|
DEAD box helicase 47 |
| chr19_-_6168518 | 0.26 |
ENSMUST00000113533.3
|
Sac3d1
|
SAC3 domain containing 1 |
| chr17_+_34416707 | 0.26 |
ENSMUST00000025196.9
|
Psmb8
|
proteasome (prosome, macropain) subunit, beta type 8 (large multifunctional peptidase 7) |
| chr15_+_81912312 | 0.25 |
ENSMUST00000230729.2
|
Xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr19_+_4036562 | 0.25 |
ENSMUST00000236224.2
ENSMUST00000236510.2 ENSMUST00000237910.2 ENSMUST00000235612.2 ENSMUST00000054030.8 |
Acy3
|
aspartoacylase (aminoacylase) 3 |
| chr7_-_126817475 | 0.25 |
ENSMUST00000106313.8
ENSMUST00000142356.3 |
Septin1
|
septin 1 |
| chr7_+_127345909 | 0.25 |
ENSMUST00000033081.14
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chrX_-_7471613 | 0.24 |
ENSMUST00000033483.5
|
Ccdc22
|
coiled-coil domain containing 22 |
| chr3_+_10077608 | 0.24 |
ENSMUST00000029046.9
|
Fabp5
|
fatty acid binding protein 5, epidermal |
| chr17_+_26895344 | 0.24 |
ENSMUST00000015719.16
|
Atp6v0e
|
ATPase, H+ transporting, lysosomal V0 subunit E |
| chr10_-_111833138 | 0.23 |
ENSMUST00000074805.12
|
Glipr1
|
GLI pathogenesis-related 1 (glioma) |
| chr12_+_112586501 | 0.23 |
ENSMUST00000180015.9
|
Adssl1
|
adenylosuccinate synthetase like 1 |
| chr11_-_84807164 | 0.23 |
ENSMUST00000103195.5
|
Znhit3
|
zinc finger, HIT type 3 |
| chr17_+_46957151 | 0.23 |
ENSMUST00000002844.14
ENSMUST00000113429.8 ENSMUST00000113430.2 |
Mrpl2
|
mitochondrial ribosomal protein L2 |
| chrX_-_106446928 | 0.23 |
ENSMUST00000033591.6
|
Itm2a
|
integral membrane protein 2A |
| chr7_+_131162338 | 0.23 |
ENSMUST00000208571.2
|
Bub3
|
BUB3 mitotic checkpoint protein |
| chr19_+_24853039 | 0.23 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr19_+_8713156 | 0.23 |
ENSMUST00000210512.2
ENSMUST00000049424.11 |
Wdr74
|
WD repeat domain 74 |
| chr4_-_136626073 | 0.23 |
ENSMUST00000046285.6
|
C1qa
|
complement component 1, q subcomponent, alpha polypeptide |
| chr2_+_103800459 | 0.23 |
ENSMUST00000111143.8
ENSMUST00000138815.2 |
Lmo2
|
LIM domain only 2 |
| chr4_-_123033721 | 0.23 |
ENSMUST00000030404.5
|
Ppie
|
peptidylprolyl isomerase E (cyclophilin E) |
| chr6_+_47930324 | 0.22 |
ENSMUST00000101445.11
|
Zfp956
|
zinc finger protein 956 |
| chr10_-_17898938 | 0.22 |
ENSMUST00000220110.2
|
Abracl
|
ABRA C-terminal like |
| chr7_-_118304930 | 0.22 |
ENSMUST00000207323.2
ENSMUST00000038791.15 |
Gde1
|
glycerophosphodiester phosphodiesterase 1 |
| chr11_-_72441054 | 0.22 |
ENSMUST00000021154.7
|
Spns3
|
spinster homolog 3 |
| chr8_-_106665060 | 0.22 |
ENSMUST00000034369.10
|
Psmb10
|
proteasome (prosome, macropain) subunit, beta type 10 |
| chrX_-_149879501 | 0.22 |
ENSMUST00000112683.9
ENSMUST00000026295.10 |
Tsr2
|
TSR2 20S rRNA accumulation |
| chr6_-_126916487 | 0.22 |
ENSMUST00000144954.5
ENSMUST00000112220.8 ENSMUST00000112221.8 |
Rad51ap1
|
RAD51 associated protein 1 |
| chr7_-_126391388 | 0.22 |
ENSMUST00000206570.2
|
Ppp4c
|
protein phosphatase 4, catalytic subunit |
| chr13_+_73479101 | 0.22 |
ENSMUST00000022098.10
ENSMUST00000222030.2 |
Mrpl36
|
mitochondrial ribosomal protein L36 |
| chr4_-_4138817 | 0.21 |
ENSMUST00000133567.2
|
Penk
|
preproenkephalin |
| chr13_-_25121568 | 0.21 |
ENSMUST00000037615.7
|
Aldh5a1
|
aldhehyde dehydrogenase family 5, subfamily A1 |
| chr3_+_103875574 | 0.21 |
ENSMUST00000063717.14
ENSMUST00000055425.15 ENSMUST00000123611.8 ENSMUST00000090685.11 |
Phtf1
|
putative homeodomain transcription factor 1 |
| chr19_-_32443978 | 0.21 |
ENSMUST00000078034.5
|
Rpl9-ps6
|
ribosomal protein L9, pseudogene 6 |
| chr6_-_24527545 | 0.21 |
ENSMUST00000118558.5
|
Ndufa5
|
NADH:ubiquinone oxidoreductase subunit A5 |
| chr9_+_7184514 | 0.21 |
ENSMUST00000215683.2
ENSMUST00000034499.10 |
Dcun1d5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
| chr13_-_32522548 | 0.21 |
ENSMUST00000041859.9
|
Gmds
|
GDP-mannose 4, 6-dehydratase |
| chr16_+_38167352 | 0.20 |
ENSMUST00000050273.9
ENSMUST00000120495.2 ENSMUST00000119704.2 |
Cox17
Gm21987
|
cytochrome c oxidase assembly protein 17, copper chaperone predicted gene 21987 |
| chr1_-_184578057 | 0.20 |
ENSMUST00000068725.10
|
Mtarc2
|
mitochondrial amidoxime reducing component 2 |
| chr9_+_113641615 | 0.20 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr2_+_157209506 | 0.20 |
ENSMUST00000081202.6
|
Manbal
|
mannosidase, beta A, lysosomal-like |
| chr6_+_18848570 | 0.20 |
ENSMUST00000056398.11
|
Lsm8
|
LSM8 homolog, U6 small nuclear RNA associated |
| chr5_-_69748126 | 0.20 |
ENSMUST00000166298.8
|
Gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr4_+_127137577 | 0.19 |
ENSMUST00000142029.2
|
Smim12
|
small integral membrane protein 12 |
| chr16_+_16120810 | 0.19 |
ENSMUST00000159962.8
ENSMUST00000059955.15 |
Yars2
|
tyrosyl-tRNA synthetase 2 (mitochondrial) |
| chr15_-_35938155 | 0.19 |
ENSMUST00000156915.3
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chr17_+_80531870 | 0.19 |
ENSMUST00000069486.13
|
Gemin6
|
gem nuclear organelle associated protein 6 |
| chr4_+_98812144 | 0.19 |
ENSMUST00000169053.2
|
Usp1
|
ubiquitin specific peptidase 1 |
| chr12_+_112586465 | 0.19 |
ENSMUST00000021726.8
|
Adssl1
|
adenylosuccinate synthetase like 1 |
| chr10_+_77449543 | 0.19 |
ENSMUST00000124024.3
|
Sumo3
|
small ubiquitin-like modifier 3 |
| chr6_+_67838100 | 0.19 |
ENSMUST00000200586.2
ENSMUST00000103309.3 |
Igkv17-127
|
immunoglobulin kappa variable 17-127 |
| chr11_+_58062467 | 0.19 |
ENSMUST00000020820.2
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chr10_+_88566918 | 0.19 |
ENSMUST00000116234.9
|
Arl1
|
ADP-ribosylation factor-like 1 |
| chr9_-_103097022 | 0.19 |
ENSMUST00000168142.8
|
Trf
|
transferrin |
| chr5_+_145060489 | 0.19 |
ENSMUST00000136074.2
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr17_+_56316305 | 0.18 |
ENSMUST00000159340.8
|
Mpnd
|
MPN domain containing |
| chr11_+_59839032 | 0.18 |
ENSMUST00000081980.7
|
Med9
|
mediator complex subunit 9 |
| chr7_-_28649094 | 0.18 |
ENSMUST00000148196.3
|
Actn4
|
actinin alpha 4 |
| chr19_-_4175837 | 0.18 |
ENSMUST00000121402.2
ENSMUST00000117831.8 |
Aip
|
aryl-hydrocarbon receptor-interacting protein |
| chr5_-_45607463 | 0.18 |
ENSMUST00000197946.5
ENSMUST00000127562.3 |
Qdpr
|
quinoid dihydropteridine reductase |
| chr11_-_3321307 | 0.18 |
ENSMUST00000101640.10
ENSMUST00000101642.10 |
Limk2
|
LIM motif-containing protein kinase 2 |
| chr4_+_130253925 | 0.18 |
ENSMUST00000105994.4
|
Snrnp40
|
small nuclear ribonucleoprotein 40 (U5) |
| chr7_-_140993967 | 0.18 |
ENSMUST00000106008.3
|
Gatd1
|
glutamine amidotransferase like class 1 domain containing 1 |
| chr2_-_174280811 | 0.18 |
ENSMUST00000016400.9
|
Ctsz
|
cathepsin Z |
| chr6_-_68713748 | 0.18 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chr10_+_82669785 | 0.18 |
ENSMUST00000219368.3
|
Txnrd1
|
thioredoxin reductase 1 |
| chrX_-_72759748 | 0.18 |
ENSMUST00000002091.6
|
Bcap31
|
B cell receptor associated protein 31 |
| chr8_+_57964956 | 0.18 |
ENSMUST00000210871.2
|
Hmgb2
|
high mobility group box 2 |
| chr3_-_105861497 | 0.17 |
ENSMUST00000199311.2
|
Atp5pb
|
ATP synthase peripheral stalk-membrane subunit b |
| chr19_-_10533088 | 0.17 |
ENSMUST00000059582.9
ENSMUST00000154383.2 |
Tmem216
|
transmembrane protein 216 |
| chr10_+_7543260 | 0.17 |
ENSMUST00000040135.9
|
Nup43
|
nucleoporin 43 |
| chr2_+_103800553 | 0.17 |
ENSMUST00000111140.3
ENSMUST00000111139.3 |
Lmo2
|
LIM domain only 2 |
| chr4_-_6454262 | 0.17 |
ENSMUST00000029910.12
|
Nsmaf
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr13_-_4659120 | 0.17 |
ENSMUST00000091848.7
ENSMUST00000110691.10 |
Akr1e1
|
aldo-keto reductase family 1, member E1 |
| chr2_+_179720416 | 0.17 |
ENSMUST00000087563.7
|
Mtg2
|
mitochondrial ribosome associated GTPase 2 |
| chr5_-_45607554 | 0.17 |
ENSMUST00000015950.12
|
Qdpr
|
quinoid dihydropteridine reductase |
| chr7_+_43000838 | 0.17 |
ENSMUST00000206299.2
ENSMUST00000121494.2 |
Siglecf
|
sialic acid binding Ig-like lectin F |
| chr15_+_78810919 | 0.17 |
ENSMUST00000089377.6
|
Lgals1
|
lectin, galactose binding, soluble 1 |
| chr2_-_32321116 | 0.17 |
ENSMUST00000127961.3
ENSMUST00000136361.8 ENSMUST00000052119.14 |
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr6_+_58810674 | 0.17 |
ENSMUST00000041401.11
|
Herc3
|
hect domain and RLD 3 |
| chr7_-_4815542 | 0.17 |
ENSMUST00000079496.9
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr2_-_32314017 | 0.17 |
ENSMUST00000113307.9
|
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr16_+_20430335 | 0.17 |
ENSMUST00000115522.10
ENSMUST00000119224.8 ENSMUST00000120394.8 ENSMUST00000079600.12 |
Eef1akmt4
Gm49333
|
EEF1A lysine methyltransferase 4 predicted gene, 49333 |
| chr19_-_38032006 | 0.17 |
ENSMUST00000172095.3
ENSMUST00000041475.16 |
Myof
|
myoferlin |
| chr10_+_93376484 | 0.16 |
ENSMUST00000092215.6
|
Ccdc38
|
coiled-coil domain containing 38 |
| chr10_-_117128763 | 0.16 |
ENSMUST00000092162.7
|
Lyz1
|
lysozyme 1 |
| chr1_-_131204651 | 0.16 |
ENSMUST00000161764.8
|
Ikbke
|
inhibitor of kappaB kinase epsilon |
| chr15_-_10714653 | 0.16 |
ENSMUST00000169385.3
|
Rai14
|
retinoic acid induced 14 |
| chr8_+_3671599 | 0.16 |
ENSMUST00000207389.2
|
Pet100
|
PET100 homolog |
| chrX_-_168103266 | 0.16 |
ENSMUST00000033717.9
ENSMUST00000112115.2 |
Hccs
|
holocytochrome c synthetase |
| chr4_-_116851571 | 0.16 |
ENSMUST00000030446.15
|
Urod
|
uroporphyrinogen decarboxylase |
| chr19_-_10181243 | 0.16 |
ENSMUST00000142241.2
ENSMUST00000116542.9 ENSMUST00000025651.6 ENSMUST00000156291.2 |
Fen1
|
flap structure specific endonuclease 1 |
| chr1_-_65218217 | 0.16 |
ENSMUST00000097709.11
|
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr15_+_34082805 | 0.16 |
ENSMUST00000022865.17
|
Mtdh
|
metadherin |
| chr15_-_54953819 | 0.16 |
ENSMUST00000110231.2
ENSMUST00000023059.13 |
Dscc1
|
DNA replication and sister chromatid cohesion 1 |
| chr9_+_64080644 | 0.16 |
ENSMUST00000034966.9
|
Rpl4
|
ribosomal protein L4 |
| chr6_-_40413056 | 0.16 |
ENSMUST00000039008.10
ENSMUST00000101492.10 |
Dennd11
|
DENN domain containing 11 |
| chr18_+_32373353 | 0.15 |
ENSMUST00000025241.7
|
Ercc3
|
excision repair cross-complementing rodent repair deficiency, complementation group 3 |
| chr4_-_59438633 | 0.15 |
ENSMUST00000040166.14
ENSMUST00000107544.2 |
Susd1
|
sushi domain containing 1 |
| chr2_+_83554741 | 0.15 |
ENSMUST00000028499.11
|
Itgav
|
integrin alpha V |
| chr11_-_119190896 | 0.15 |
ENSMUST00000026667.15
|
Eif4a3
|
eukaryotic translation initiation factor 4A3 |
| chr6_-_129252323 | 0.15 |
ENSMUST00000204411.2
|
Cd69
|
CD69 antigen |
| chr17_-_26314438 | 0.15 |
ENSMUST00000236547.2
|
Nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr16_-_91443794 | 0.15 |
ENSMUST00000232367.2
ENSMUST00000231380.2 ENSMUST00000231444.2 ENSMUST00000232289.2 ENSMUST00000120450.2 ENSMUST00000023684.14 |
Gart
|
phosphoribosylglycinamide formyltransferase |
| chr12_-_55061117 | 0.15 |
ENSMUST00000172875.8
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr16_-_21606546 | 0.15 |
ENSMUST00000023559.7
|
Ehhadh
|
enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase |
| chr10_-_62285458 | 0.15 |
ENSMUST00000020273.16
|
Supv3l1
|
suppressor of var1, 3-like 1 (S. cerevisiae) |
| chr4_-_116851550 | 0.15 |
ENSMUST00000130273.8
|
Urod
|
uroporphyrinogen decarboxylase |
| chr2_+_26209755 | 0.15 |
ENSMUST00000066889.13
|
Gpsm1
|
G-protein signalling modulator 1 (AGS3-like, C. elegans) |
| chr16_+_32877382 | 0.15 |
ENSMUST00000115075.2
|
Rpl35a
|
ribosomal protein L35A |
| chr5_-_23988696 | 0.15 |
ENSMUST00000119946.8
|
Pus7
|
pseudouridylate synthase 7 |
| chrX_-_101232978 | 0.15 |
ENSMUST00000033683.8
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr15_-_11399666 | 0.15 |
ENSMUST00000022849.7
|
Tars
|
threonyl-tRNA synthetase |
| chr2_+_26800757 | 0.15 |
ENSMUST00000102898.5
|
Rpl7a
|
ribosomal protein L7A |
| chrX_-_100312629 | 0.14 |
ENSMUST00000117736.2
|
Gm20489
|
predicted gene 20489 |
| chr8_-_25506756 | 0.14 |
ENSMUST00000084032.6
ENSMUST00000207132.2 |
Adam9
|
a disintegrin and metallopeptidase domain 9 (meltrin gamma) |
| chr17_-_26314461 | 0.14 |
ENSMUST00000236128.2
ENSMUST00000025007.7 |
Nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr3_+_67490068 | 0.14 |
ENSMUST00000029344.10
|
Mfsd1
|
major facilitator superfamily domain containing 1 |
| chr9_-_36678868 | 0.14 |
ENSMUST00000217599.2
ENSMUST00000120381.9 |
Stt3a
|
STT3, subunit of the oligosaccharyltransferase complex, homolog A (S. cerevisiae) |
| chr8_-_27618643 | 0.14 |
ENSMUST00000033877.6
|
Brf2
|
BRF2, RNA polymerase III transcription initiation factor 50kDa subunit |
| chr3_+_67799510 | 0.14 |
ENSMUST00000063263.5
ENSMUST00000182006.4 |
Iqcj
Iqschfp
|
IQ motif containing J Iqcj and Schip1 fusion protein |
| chr15_+_65682066 | 0.14 |
ENSMUST00000211878.2
|
Efr3a
|
EFR3 homolog A |
| chr6_-_24528012 | 0.14 |
ENSMUST00000023851.9
|
Ndufa5
|
NADH:ubiquinone oxidoreductase subunit A5 |
| chr18_-_75094323 | 0.14 |
ENSMUST00000066532.5
|
Lipg
|
lipase, endothelial |
| chr11_-_86148344 | 0.14 |
ENSMUST00000136469.2
ENSMUST00000018212.13 |
Ints2
|
integrator complex subunit 2 |
| chr19_-_38031774 | 0.14 |
ENSMUST00000226068.2
|
Myof
|
myoferlin |
| chr10_-_117628565 | 0.14 |
ENSMUST00000167943.8
ENSMUST00000064848.7 |
Nup107
|
nucleoporin 107 |
| chr17_+_28426831 | 0.14 |
ENSMUST00000233264.2
|
Def6
|
differentially expressed in FDCP 6 |
| chr1_-_63215812 | 0.14 |
ENSMUST00000185847.2
ENSMUST00000185732.7 ENSMUST00000188370.7 ENSMUST00000168099.9 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr7_+_130179063 | 0.14 |
ENSMUST00000207918.2
ENSMUST00000215492.2 ENSMUST00000084513.12 ENSMUST00000059145.14 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr2_+_29692638 | 0.14 |
ENSMUST00000080065.3
|
Slc27a4
|
solute carrier family 27 (fatty acid transporter), member 4 |
| chr17_+_34149820 | 0.14 |
ENSMUST00000234226.2
|
Rgl2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr17_-_74623154 | 0.14 |
ENSMUST00000232687.2
|
Dpy30
|
dpy-30, histone methyltransferase complex regulatory subunit |
| chr2_-_30720345 | 0.13 |
ENSMUST00000041726.4
|
Asb6
|
ankyrin repeat and SOCS box-containing 6 |
| chr10_+_128626772 | 0.13 |
ENSMUST00000219404.2
ENSMUST00000026411.8 |
Mmp19
|
matrix metallopeptidase 19 |
| chr11_+_60345412 | 0.13 |
ENSMUST00000018568.4
|
Drg2
|
developmentally regulated GTP binding protein 2 |
| chr13_+_55747868 | 0.13 |
ENSMUST00000133176.8
ENSMUST00000064701.8 |
B4galt7
|
xylosylprotein beta1,4-galactosyltransferase, polypeptide 7 (galactosyltransferase I) |
| chr9_-_103099262 | 0.13 |
ENSMUST00000170904.2
|
Trf
|
transferrin |
| chr6_+_29859372 | 0.13 |
ENSMUST00000115238.10
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr2_-_174305856 | 0.13 |
ENSMUST00000016396.8
|
Atp5e
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
| chr9_+_21634779 | 0.13 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr9_+_21504018 | 0.13 |
ENSMUST00000062125.11
|
Timm29
|
translocase of inner mitochondrial membrane 29 |
| chrX_-_141173330 | 0.13 |
ENSMUST00000112907.8
|
Acsl4
|
acyl-CoA synthetase long-chain family member 4 |
| chr12_+_69771713 | 0.13 |
ENSMUST00000220916.2
ENSMUST00000021372.6 ENSMUST00000220539.2 ENSMUST00000220460.2 |
Dmac2l
|
distal membrane arm assembly complex 2 like |
| chr2_+_61542038 | 0.13 |
ENSMUST00000028278.14
|
Psmd14
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 |
| chr3_-_19319155 | 0.13 |
ENSMUST00000091314.11
|
Pde7a
|
phosphodiesterase 7A |
| chr4_+_98812047 | 0.13 |
ENSMUST00000030289.9
|
Usp1
|
ubiquitin specific peptidase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ubp1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 1.3 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.4 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.3 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.1 | 0.4 | GO:2000984 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.1 | 0.3 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.1 | 0.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.3 | GO:0071727 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 0.3 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.1 | 0.2 | GO:1904959 | regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.1 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.3 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.1 | 0.4 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 0.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 0.4 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 0.4 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.1 | GO:1900756 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.1 | 0.3 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.5 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.0 | 0.2 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.3 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.2 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.0 | 0.3 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.1 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.2 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.2 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.1 | GO:0002590 | negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.0 | 0.7 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.3 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.2 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.2 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.4 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 0.3 | GO:0061502 | uropod organization(GO:0032796) early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.1 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.2 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.3 | GO:0019660 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.1 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.2 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.3 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.2 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.1 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 0.0 | 0.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:2000471 | immunoglobulin biosynthetic process(GO:0002378) regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.0 | 0.1 | GO:0010138 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) CMP metabolic process(GO:0046035) |
| 0.0 | 0.3 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.0 | 0.3 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:2000850 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.6 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.0 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.3 | GO:1904867 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.1 | GO:1903898 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.2 | GO:0071554 | cell wall macromolecule catabolic process(GO:0016998) cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.0 | 0.1 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 0.0 | GO:0002414 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.0 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.2 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.1 | GO:1904180 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.0 | GO:0071332 | oxalic acid secretion(GO:0046724) cellular response to fructose stimulus(GO:0071332) |
| 0.0 | 0.0 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.1 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.3 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.5 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.0 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.2 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.2 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.0 | 0.1 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.4 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.1 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.0 | 0.0 | GO:0038091 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.0 | 0.1 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.0 | 0.0 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.0 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.0 | 0.1 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.4 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.0 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 0.0 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.0 | 0.2 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.1 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.3 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.3 | GO:0044753 | amphisome(GO:0044753) |
| 0.1 | 0.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.3 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.1 | 0.2 | GO:1903754 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.1 | 0.2 | GO:0033597 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.1 | 0.2 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.2 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.2 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.0 | 0.1 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.3 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.6 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.2 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.5 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 0.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 1.1 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 1.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.0 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.4 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 0.3 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.1 | 0.3 | GO:0035730 | S-nitrosoglutathione binding(GO:0035730) dinitrosyl-iron complex binding(GO:0035731) |
| 0.1 | 0.3 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 0.3 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.1 | 0.2 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.1 | 0.2 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 0.2 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.1 | 0.3 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 0.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.2 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.3 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.3 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.3 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.0 | 0.1 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.2 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 0.0 | 0.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.2 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.1 | GO:0070401 | NADP+ binding(GO:0070401) |
| 0.0 | 0.1 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.3 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.5 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.2 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.3 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.3 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.4 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.4 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.0 | 0.1 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.0 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.3 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.7 | PID REELIN PATHWAY | Reelin signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 2.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.0 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.5 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.3 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.0 | 0.2 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |