Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Xbp1_Creb3l1
Z-value: 1.75
Transcription factors associated with Xbp1_Creb3l1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
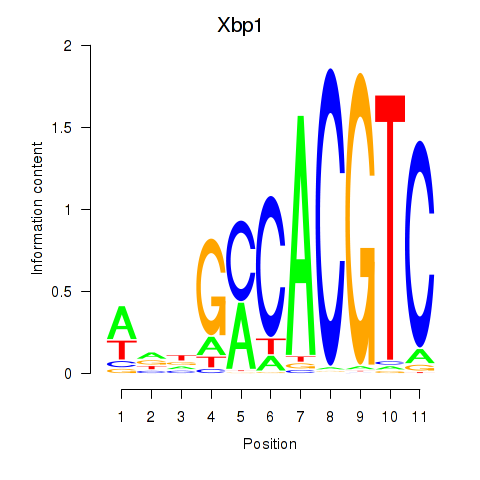
Xbp1
|
ENSMUSG00000020484.20 | X-box binding protein 1 |
|
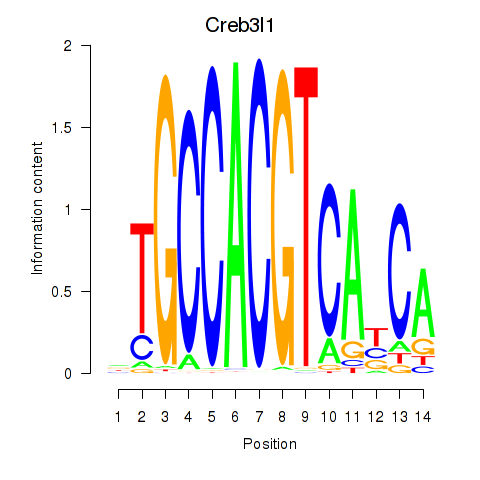
Creb3l1
|
ENSMUSG00000027230.10 | cAMP responsive element binding protein 3-like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Creb3l1 | mm39_v1_chr2_-_91854844_91854847 | 0.35 | 5.6e-01 | Click! |
| Xbp1 | mm39_v1_chr11_+_5470652_5470659 | 0.23 | 7.1e-01 | Click! |
Activity profile of Xbp1_Creb3l1 motif
Sorted Z-values of Xbp1_Creb3l1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_72935382 | 2.13 |
ENSMUST00000144337.2
|
Tmsb10
|
thymosin, beta 10 |
| chr3_+_123061094 | 1.35 |
ENSMUST00000047923.12
ENSMUST00000200333.2 |
Sec24d
|
Sec24 related gene family, member D (S. cerevisiae) |
| chr15_+_31224555 | 1.17 |
ENSMUST00000186109.2
|
Dap
|
death-associated protein |
| chr6_-_72935468 | 1.06 |
ENSMUST00000114050.8
|
Tmsb10
|
thymosin, beta 10 |
| chr6_-_72935171 | 0.96 |
ENSMUST00000114049.2
|
Tmsb10
|
thymosin, beta 10 |
| chr15_+_31224616 | 0.95 |
ENSMUST00000186547.7
|
Dap
|
death-associated protein |
| chr14_+_26359191 | 0.91 |
ENSMUST00000022429.9
|
Arf4
|
ADP-ribosylation factor 4 |
| chr14_+_26359390 | 0.88 |
ENSMUST00000112318.10
|
Arf4
|
ADP-ribosylation factor 4 |
| chr3_-_131196213 | 0.83 |
ENSMUST00000197057.2
|
Sgms2
|
sphingomyelin synthase 2 |
| chr2_+_80145805 | 0.82 |
ENSMUST00000028392.8
|
Dnajc10
|
DnaJ heat shock protein family (Hsp40) member C10 |
| chr15_+_31224460 | 0.76 |
ENSMUST00000044524.16
|
Dap
|
death-associated protein |
| chr2_+_70491901 | 0.74 |
ENSMUST00000112201.8
ENSMUST00000028509.11 ENSMUST00000133432.8 ENSMUST00000112205.2 |
Gorasp2
|
golgi reassembly stacking protein 2 |
| chr8_+_26210064 | 0.70 |
ENSMUST00000068916.16
ENSMUST00000139836.8 |
Plpp5
|
phospholipid phosphatase 5 |
| chr10_+_128744689 | 0.66 |
ENSMUST00000105229.9
|
Cd63
|
CD63 antigen |
| chr19_+_32573182 | 0.65 |
ENSMUST00000235594.2
|
Papss2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr6_-_52181393 | 0.64 |
ENSMUST00000048794.7
|
Hoxa5
|
homeobox A5 |
| chr9_-_89586960 | 0.56 |
ENSMUST00000058488.9
|
Tmed3
|
transmembrane p24 trafficking protein 3 |
| chr10_-_18619439 | 0.48 |
ENSMUST00000019999.7
|
Arfgef3
|
ARFGEF family member 3 |
| chr5_-_148989821 | 0.48 |
ENSMUST00000139443.8
ENSMUST00000085546.13 |
Hmgb1
|
high mobility group box 1 |
| chr9_+_86454018 | 0.47 |
ENSMUST00000185566.7
ENSMUST00000034988.10 ENSMUST00000179212.3 |
Rwdd2a
|
RWD domain containing 2A |
| chr18_+_57487908 | 0.45 |
ENSMUST00000238069.2
|
Prrc1
|
proline-rich coiled-coil 1 |
| chr2_+_164339438 | 0.42 |
ENSMUST00000103101.11
ENSMUST00000117066.8 |
Pigt
|
phosphatidylinositol glycan anchor biosynthesis, class T |
| chr13_-_38712387 | 0.39 |
ENSMUST00000035988.16
|
Txndc5
|
thioredoxin domain containing 5 |
| chr19_-_5821442 | 0.39 |
ENSMUST00000236978.2
|
Scyl1
|
SCY1-like 1 (S. cerevisiae) |
| chr3_-_27764571 | 0.38 |
ENSMUST00000046157.10
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr7_+_116103599 | 0.38 |
ENSMUST00000032895.15
|
Nucb2
|
nucleobindin 2 |
| chr7_+_116103643 | 0.37 |
ENSMUST00000183175.8
|
Nucb2
|
nucleobindin 2 |
| chr10_-_84369831 | 0.36 |
ENSMUST00000167671.2
ENSMUST00000053871.5 |
Ckap4
|
cytoskeleton-associated protein 4 |
| chr6_+_29348068 | 0.35 |
ENSMUST00000173216.8
ENSMUST00000173694.5 ENSMUST00000172974.8 ENSMUST00000031779.17 ENSMUST00000090481.14 |
Calu
|
calumenin |
| chr10_-_18619658 | 0.35 |
ENSMUST00000215836.2
|
Arfgef3
|
ARFGEF family member 3 |
| chr19_-_5821373 | 0.32 |
ENSMUST00000236297.2
ENSMUST00000236773.2 ENSMUST00000025890.10 |
Scyl1
|
SCY1-like 1 (S. cerevisiae) |
| chr4_+_47474652 | 0.32 |
ENSMUST00000065678.6
|
Sec61b
|
Sec61 beta subunit |
| chr1_-_13660027 | 0.30 |
ENSMUST00000027068.11
|
Tram1
|
translocating chain-associating membrane protein 1 |
| chr1_+_157334347 | 0.30 |
ENSMUST00000027881.15
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr1_+_157334298 | 0.30 |
ENSMUST00000086130.9
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr12_-_69230760 | 0.29 |
ENSMUST00000110620.2
ENSMUST00000110619.2 ENSMUST00000054544.7 |
Rpl36al
|
ribosomal protein L36A-like |
| chr7_+_26958150 | 0.28 |
ENSMUST00000079258.7
|
Numbl
|
numb-like |
| chr8_+_70755168 | 0.27 |
ENSMUST00000066469.14
|
Cope
|
coatomer protein complex, subunit epsilon |
| chr4_+_47474715 | 0.27 |
ENSMUST00000137461.8
ENSMUST00000125622.2 |
Sec61b
|
Sec61 beta subunit |
| chr1_-_183150867 | 0.26 |
ENSMUST00000194543.4
|
Mia3
|
melanoma inhibitory activity 3 |
| chr19_-_6134703 | 0.26 |
ENSMUST00000161548.8
|
Zfpl1
|
zinc finger like protein 1 |
| chr4_+_33031342 | 0.26 |
ENSMUST00000124992.8
|
Ube2j1
|
ubiquitin-conjugating enzyme E2J 1 |
| chr11_+_3464861 | 0.25 |
ENSMUST00000094469.6
|
Selenom
|
selenoprotein M |
| chr17_+_34341766 | 0.24 |
ENSMUST00000042121.11
|
H2-DMa
|
histocompatibility 2, class II, locus DMa |
| chr10_+_128745214 | 0.23 |
ENSMUST00000220308.2
|
Cd63
|
CD63 antigen |
| chr16_-_35311243 | 0.23 |
ENSMUST00000023550.9
|
Pdia5
|
protein disulfide isomerase associated 5 |
| chr17_-_34250474 | 0.21 |
ENSMUST00000171872.3
ENSMUST00000025186.16 |
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr7_+_128125339 | 0.21 |
ENSMUST00000033136.9
|
Bag3
|
BCL2-associated athanogene 3 |
| chrX_+_72830668 | 0.20 |
ENSMUST00000002090.3
|
Ssr4
|
signal sequence receptor, delta |
| chr19_-_6886965 | 0.20 |
ENSMUST00000173091.2
|
Prdx5
|
peroxiredoxin 5 |
| chr6_-_88495835 | 0.19 |
ENSMUST00000032168.7
|
Sec61a1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr1_-_171910324 | 0.19 |
ENSMUST00000003550.11
|
Ncstn
|
nicastrin |
| chr19_+_46750016 | 0.18 |
ENSMUST00000099373.12
ENSMUST00000077666.6 |
Cnnm2
|
cyclin M2 |
| chr9_-_21003268 | 0.18 |
ENSMUST00000115487.3
|
Raver1
|
ribonucleoprotein, PTB-binding 1 |
| chrX_+_72830607 | 0.17 |
ENSMUST00000166518.8
|
Ssr4
|
signal sequence receptor, delta |
| chr7_-_44741622 | 0.17 |
ENSMUST00000210469.2
ENSMUST00000211352.2 ENSMUST00000019683.11 |
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr10_-_67120959 | 0.16 |
ENSMUST00000159002.2
ENSMUST00000077839.13 |
Nrbf2
|
nuclear receptor binding factor 2 |
| chr11_-_86698484 | 0.16 |
ENSMUST00000018569.14
|
Dhx40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr6_-_4086914 | 0.16 |
ENSMUST00000049166.5
|
Bet1
|
Bet1 golgi vesicular membrane trafficking protein |
| chr7_+_79910948 | 0.16 |
ENSMUST00000117989.2
|
Ngrn
|
neugrin, neurite outgrowth associated |
| chr17_-_56490887 | 0.16 |
ENSMUST00000019723.8
|
Mydgf
|
myeloid derived growth factor |
| chr15_-_57755753 | 0.15 |
ENSMUST00000022993.7
|
Derl1
|
Der1-like domain family, member 1 |
| chr7_+_49559859 | 0.15 |
ENSMUST00000056442.12
|
Slc6a5
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 5 |
| chr3_+_89153704 | 0.15 |
ENSMUST00000168900.3
|
Krtcap2
|
keratinocyte associated protein 2 |
| chr11_+_53991750 | 0.15 |
ENSMUST00000093107.12
ENSMUST00000019050.12 ENSMUST00000174616.8 ENSMUST00000129499.8 ENSMUST00000126840.8 |
P4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha II polypeptide |
| chr6_+_88061464 | 0.15 |
ENSMUST00000032143.8
|
Rpn1
|
ribophorin I |
| chr5_+_113873864 | 0.15 |
ENSMUST00000065698.7
|
Ficd
|
FIC domain containing |
| chrX_-_72830487 | 0.15 |
ENSMUST00000052761.9
|
Idh3g
|
isocitrate dehydrogenase 3 (NAD+), gamma |
| chr13_-_76246689 | 0.14 |
ENSMUST00000239063.2
ENSMUST00000120573.3 |
Arsk
|
arylsulfatase K |
| chr19_+_32734884 | 0.14 |
ENSMUST00000013807.8
|
Pten
|
phosphatase and tensin homolog |
| chr10_+_41395410 | 0.14 |
ENSMUST00000019962.15
|
Cd164
|
CD164 antigen |
| chr15_+_79575046 | 0.14 |
ENSMUST00000046463.10
|
Gtpbp1
|
GTP binding protein 1 |
| chr11_-_20781009 | 0.14 |
ENSMUST00000047028.9
|
Lgalsl
|
lectin, galactoside binding-like |
| chr3_+_144276448 | 0.13 |
ENSMUST00000106211.3
|
Selenof
|
selenoprotein F |
| chr9_+_108216466 | 0.13 |
ENSMUST00000193987.2
|
Gpx1
|
glutathione peroxidase 1 |
| chr12_+_59177552 | 0.13 |
ENSMUST00000175877.8
|
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr12_+_51424343 | 0.13 |
ENSMUST00000219434.2
ENSMUST00000021335.7 |
Scfd1
|
Sec1 family domain containing 1 |
| chr1_-_10038030 | 0.12 |
ENSMUST00000185184.2
|
Tcf24
|
transcription factor 24 |
| chr7_+_138792890 | 0.12 |
ENSMUST00000016124.15
|
Lrrc27
|
leucine rich repeat containing 27 |
| chr19_+_6887059 | 0.12 |
ENSMUST00000088257.14
ENSMUST00000116551.10 |
Trmt112
|
tRNA methyltransferase 11-2 |
| chr11_-_93776580 | 0.12 |
ENSMUST00000066888.10
|
Utp18
|
UTP18 small subunit processome component |
| chr17_-_26004298 | 0.11 |
ENSMUST00000150324.8
|
Haghl
|
hydroxyacylglutathione hydrolase-like |
| chr3_+_144276154 | 0.11 |
ENSMUST00000082437.11
|
Selenof
|
selenoprotein F |
| chr19_-_6886898 | 0.11 |
ENSMUST00000238095.2
|
Prdx5
|
peroxiredoxin 5 |
| chr19_+_5138562 | 0.11 |
ENSMUST00000238093.2
ENSMUST00000025811.6 ENSMUST00000237025.2 |
Yif1a
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr5_+_77122530 | 0.10 |
ENSMUST00000101087.10
ENSMUST00000120550.2 |
Srp72
|
signal recognition particle 72 |
| chr18_+_34464185 | 0.10 |
ENSMUST00000072576.10
ENSMUST00000119329.8 |
Srp19
|
signal recognition particle 19 |
| chr7_+_45434755 | 0.10 |
ENSMUST00000233503.2
ENSMUST00000120005.10 ENSMUST00000211609.2 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chr8_-_70755132 | 0.10 |
ENSMUST00000008004.10
|
Ddx49
|
DEAD box helicase 49 |
| chr6_-_29212295 | 0.10 |
ENSMUST00000078155.12
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chr4_-_57300361 | 0.10 |
ENSMUST00000153926.8
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr3_-_84489923 | 0.10 |
ENSMUST00000143514.3
|
Arfip1
|
ADP-ribosylation factor interacting protein 1 |
| chr5_+_65127412 | 0.09 |
ENSMUST00000031080.15
|
Fam114a1
|
family with sequence similarity 114, member A1 |
| chr17_-_34250616 | 0.09 |
ENSMUST00000169397.9
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr4_+_154041687 | 0.09 |
ENSMUST00000105645.9
ENSMUST00000058393.9 ENSMUST00000141493.8 |
A430005L14Rik
|
RIKEN cDNA A430005L14 gene |
| chr10_-_84938350 | 0.09 |
ENSMUST00000059383.8
ENSMUST00000216889.2 |
Fhl4
|
four and a half LIM domains 4 |
| chr19_-_5345423 | 0.09 |
ENSMUST00000235182.2
ENSMUST00000025774.11 |
Sf3b2
|
splicing factor 3b, subunit 2 |
| chr9_-_107109108 | 0.09 |
ENSMUST00000044532.11
|
Dock3
|
dedicator of cyto-kinesis 3 |
| chr19_+_6326755 | 0.09 |
ENSMUST00000025684.4
|
Ehd1
|
EH-domain containing 1 |
| chr4_+_148675939 | 0.09 |
ENSMUST00000006611.9
|
Srm
|
spermidine synthase |
| chr17_-_83821716 | 0.09 |
ENSMUST00000025095.9
ENSMUST00000167741.9 |
Cox7a2l
|
cytochrome c oxidase subunit 7A2 like |
| chr10_-_83173708 | 0.08 |
ENSMUST00000039956.6
|
Slc41a2
|
solute carrier family 41, member 2 |
| chr4_-_151946124 | 0.08 |
ENSMUST00000169423.9
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr10_+_41395870 | 0.08 |
ENSMUST00000189300.2
|
Cd164
|
CD164 antigen |
| chr9_+_108216433 | 0.08 |
ENSMUST00000191997.2
|
Gpx1
|
glutathione peroxidase 1 |
| chr4_-_43562397 | 0.08 |
ENSMUST00000030187.14
|
Tln1
|
talin 1 |
| chr2_-_58247764 | 0.08 |
ENSMUST00000112608.9
ENSMUST00000112607.3 ENSMUST00000028178.14 |
Acvr1c
|
activin A receptor, type IC |
| chr9_+_55056648 | 0.07 |
ENSMUST00000121677.8
|
Ube2q2
|
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr2_-_168072493 | 0.07 |
ENSMUST00000109193.8
|
Dpm1
|
dolichol-phosphate (beta-D) mannosyltransferase 1 |
| chr9_+_44290787 | 0.07 |
ENSMUST00000066601.13
|
Hyou1
|
hypoxia up-regulated 1 |
| chr17_+_53786240 | 0.07 |
ENSMUST00000017975.7
|
Rab5a
|
RAB5A, member RAS oncogene family |
| chr10_-_121933276 | 0.07 |
ENSMUST00000140299.3
|
Rxylt1
|
ribitol xylosyltransferase 1 |
| chr6_-_102550062 | 0.07 |
ENSMUST00000203619.3
|
Cntn3
|
contactin 3 |
| chr18_+_52464577 | 0.07 |
ENSMUST00000025388.7
|
Ftmt
|
ferritin mitochondrial |
| chr17_-_34340918 | 0.07 |
ENSMUST00000151986.2
|
Brd2
|
bromodomain containing 2 |
| chr11_+_62139782 | 0.07 |
ENSMUST00000018644.3
|
Adora2b
|
adenosine A2b receptor |
| chr7_-_16657825 | 0.07 |
ENSMUST00000019514.10
|
Calm3
|
calmodulin 3 |
| chr11_+_94520567 | 0.07 |
ENSMUST00000021239.7
|
Lrrc59
|
leucine rich repeat containing 59 |
| chr7_-_126986331 | 0.07 |
ENSMUST00000049052.4
|
9130019O22Rik
|
RIKEN cDNA 9130019O22 gene |
| chr1_-_75455915 | 0.06 |
ENSMUST00000079205.14
ENSMUST00000094818.4 |
Chpf
|
chondroitin polymerizing factor |
| chr7_+_44813363 | 0.06 |
ENSMUST00000085374.7
ENSMUST00000209634.2 |
Slc17a7
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 |
| chr5_+_143389573 | 0.06 |
ENSMUST00000110731.4
|
Kdelr2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr7_-_44861235 | 0.06 |
ENSMUST00000210741.2
ENSMUST00000209466.2 |
Dkkl1
|
dickkopf-like 1 |
| chr5_-_72716942 | 0.06 |
ENSMUST00000074948.5
ENSMUST00000087216.12 |
Nfxl1
|
nuclear transcription factor, X-box binding-like 1 |
| chr9_+_57468217 | 0.06 |
ENSMUST00000045791.11
ENSMUST00000216986.2 |
Scamp2
|
secretory carrier membrane protein 2 |
| chr6_-_29212239 | 0.06 |
ENSMUST00000160878.8
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chr8_-_106692668 | 0.06 |
ENSMUST00000116429.9
ENSMUST00000034370.17 |
Slc12a4
|
solute carrier family 12, member 4 |
| chr1_+_75456173 | 0.06 |
ENSMUST00000113575.9
ENSMUST00000148980.2 ENSMUST00000050899.7 ENSMUST00000187411.2 |
Tmem198
|
transmembrane protein 198 |
| chr13_-_55469735 | 0.06 |
ENSMUST00000035242.9
|
Rab24
|
RAB24, member RAS oncogene family |
| chr4_+_109835224 | 0.06 |
ENSMUST00000061187.4
|
Dmrta2
|
doublesex and mab-3 related transcription factor like family A2 |
| chr2_-_143853122 | 0.05 |
ENSMUST00000016072.12
ENSMUST00000037875.6 |
Rrbp1
|
ribosome binding protein 1 |
| chr7_-_121666486 | 0.05 |
ENSMUST00000033159.4
|
Ears2
|
glutamyl-tRNA synthetase 2, mitochondrial |
| chr4_-_151946219 | 0.05 |
ENSMUST00000097774.9
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr6_-_54570124 | 0.05 |
ENSMUST00000046520.13
|
Fkbp14
|
FK506 binding protein 14 |
| chr5_+_31026967 | 0.05 |
ENSMUST00000114716.4
|
Tmem214
|
transmembrane protein 214 |
| chr4_-_155737841 | 0.05 |
ENSMUST00000030937.2
|
Mmp23
|
matrix metallopeptidase 23 |
| chr7_-_44741138 | 0.05 |
ENSMUST00000210527.2
|
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr4_+_43578922 | 0.05 |
ENSMUST00000030190.9
|
Rgp1
|
RAB6A GEF compex partner 1 |
| chr3_-_19682833 | 0.05 |
ENSMUST00000119133.2
|
1700064H15Rik
|
RIKEN cDNA 1700064H15 gene |
| chr10_+_42637479 | 0.05 |
ENSMUST00000019937.5
|
Sec63
|
SEC63-like (S. cerevisiae) |
| chr16_+_51851917 | 0.05 |
ENSMUST00000227062.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr11_-_68743944 | 0.05 |
ENSMUST00000018880.14
|
Ndel1
|
nudE neurodevelopment protein 1 like 1 |
| chr11_-_72686627 | 0.05 |
ENSMUST00000079681.6
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr2_-_168584020 | 0.05 |
ENSMUST00000109177.8
|
Atp9a
|
ATPase, class II, type 9A |
| chr16_+_51851948 | 0.05 |
ENSMUST00000226593.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr4_+_150939521 | 0.05 |
ENSMUST00000030811.2
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr2_+_153716958 | 0.04 |
ENSMUST00000028983.3
|
Bpifb2
|
BPI fold containing family B, member 2 |
| chr1_-_71454041 | 0.04 |
ENSMUST00000087268.7
|
Abca12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr7_+_138792919 | 0.04 |
ENSMUST00000156768.8
|
Lrrc27
|
leucine rich repeat containing 27 |
| chr11_-_96807273 | 0.04 |
ENSMUST00000103152.11
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr3_-_27764522 | 0.03 |
ENSMUST00000195008.6
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr16_+_17716480 | 0.03 |
ENSMUST00000055374.8
|
Tssk2
|
testis-specific serine kinase 2 |
| chr8_+_71819854 | 0.03 |
ENSMUST00000019169.8
|
Use1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr1_-_3741721 | 0.03 |
ENSMUST00000070533.5
|
Xkr4
|
X-linked Kx blood group related 4 |
| chr14_-_60324265 | 0.03 |
ENSMUST00000080368.13
|
Atp8a2
|
ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 |
| chr9_+_7571397 | 0.03 |
ENSMUST00000120900.8
ENSMUST00000151853.8 ENSMUST00000152878.3 |
Mmp27
|
matrix metallopeptidase 27 |
| chr9_+_44290832 | 0.03 |
ENSMUST00000161318.8
ENSMUST00000217019.2 ENSMUST00000160902.8 |
Hyou1
|
hypoxia up-regulated 1 |
| chr14_+_63673843 | 0.03 |
ENSMUST00000121288.2
|
Fam167a
|
family with sequence similarity 167, member A |
| chr15_-_48655329 | 0.03 |
ENSMUST00000160658.8
ENSMUST00000100670.10 ENSMUST00000162830.8 |
Csmd3
|
CUB and Sushi multiple domains 3 |
| chr8_-_46747629 | 0.03 |
ENSMUST00000058636.9
|
Helt
|
helt bHLH transcription factor |
| chr7_-_127048165 | 0.03 |
ENSMUST00000106299.2
|
Zfp689
|
zinc finger protein 689 |
| chr8_+_71819534 | 0.03 |
ENSMUST00000110054.8
ENSMUST00000139541.8 |
Use1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr9_+_8900460 | 0.03 |
ENSMUST00000070463.10
ENSMUST00000098986.4 |
Pgr
|
progesterone receptor |
| chr4_-_108263873 | 0.02 |
ENSMUST00000184609.2
|
Gpx7
|
glutathione peroxidase 7 |
| chr5_-_97540230 | 0.02 |
ENSMUST00000060265.6
|
Naa11
|
N(alpha)-acetyltransferase 11, NatA catalytic subunit |
| chr12_+_88920169 | 0.02 |
ENSMUST00000057634.14
|
Nrxn3
|
neurexin III |
| chr14_-_21798678 | 0.02 |
ENSMUST00000075040.9
|
Dusp13
|
dual specificity phosphatase 13 |
| chr19_+_3758285 | 0.02 |
ENSMUST00000237320.2
ENSMUST00000039048.2 ENSMUST00000235295.2 ENSMUST00000237955.2 ENSMUST00000235837.2 |
1810055G02Rik
|
RIKEN cDNA 1810055G02 gene |
| chr9_-_103079312 | 0.02 |
ENSMUST00000035157.10
|
Srprb
|
signal recognition particle receptor, B subunit |
| chr11_+_53992054 | 0.02 |
ENSMUST00000135653.8
|
P4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha II polypeptide |
| chr4_+_33031433 | 0.02 |
ENSMUST00000029944.13
|
Ube2j1
|
ubiquitin-conjugating enzyme E2J 1 |
| chr7_-_126975199 | 0.02 |
ENSMUST00000067425.6
|
Zfp747
|
zinc finger protein 747 |
| chr12_-_4088905 | 0.02 |
ENSMUST00000111178.2
|
Efr3b
|
EFR3 homolog B |
| chr16_+_51851588 | 0.02 |
ENSMUST00000114471.3
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr19_+_8718777 | 0.02 |
ENSMUST00000176381.8
|
Stx5a
|
syntaxin 5A |
| chr7_-_81642034 | 0.02 |
ENSMUST00000026096.10
|
Bnc1
|
basonuclin 1 |
| chr10_-_128236366 | 0.02 |
ENSMUST00000219131.2
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr6_-_126717590 | 0.02 |
ENSMUST00000185333.2
|
Kcna6
|
potassium voltage-gated channel, shaker-related, subfamily, member 6 |
| chr18_+_51250748 | 0.01 |
ENSMUST00000116639.4
|
Prr16
|
proline rich 16 |
| chr4_-_137523659 | 0.01 |
ENSMUST00000030551.11
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr2_-_6327884 | 0.01 |
ENSMUST00000238876.2
|
1700014B07Rik
|
RIKEN cDNA 1700014B07 gene |
| chr10_-_128236317 | 0.01 |
ENSMUST00000167859.2
ENSMUST00000218858.2 |
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr8_+_126722113 | 0.01 |
ENSMUST00000212831.2
|
Kcnk1
|
potassium channel, subfamily K, member 1 |
| chr15_+_23036535 | 0.01 |
ENSMUST00000164787.8
|
Cdh18
|
cadherin 18 |
| chr7_+_6392535 | 0.01 |
ENSMUST00000207465.2
ENSMUST00000208338.2 |
Zfp28
|
zinc finger protein 28 |
| chr4_-_151946155 | 0.01 |
ENSMUST00000049790.14
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr11_+_78079631 | 0.01 |
ENSMUST00000056241.12
ENSMUST00000207728.2 |
Rab34
|
RAB34, member RAS oncogene family |
| chr6_-_126717114 | 0.01 |
ENSMUST00000112242.2
|
Kcna6
|
potassium voltage-gated channel, shaker-related, subfamily, member 6 |
| chr16_-_32698091 | 0.01 |
ENSMUST00000119810.2
|
Rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr19_+_8875459 | 0.01 |
ENSMUST00000096246.5
ENSMUST00000235274.2 |
Ganab
|
alpha glucosidase 2 alpha neutral subunit |
| chr2_+_32460868 | 0.01 |
ENSMUST00000140592.8
ENSMUST00000028151.7 |
Dpm2
|
dolichol-phosphate (beta-D) mannosyltransferase 2 |
| chr8_+_126721878 | 0.01 |
ENSMUST00000046765.10
|
Kcnk1
|
potassium channel, subfamily K, member 1 |
| chr3_-_107993906 | 0.01 |
ENSMUST00000102638.8
ENSMUST00000102637.8 |
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr8_-_106863423 | 0.01 |
ENSMUST00000146940.2
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr11_+_85777185 | 0.01 |
ENSMUST00000108047.8
|
Tbx4
|
T-box 4 |
| chr14_-_55163311 | 0.01 |
ENSMUST00000022813.8
|
Efs
|
embryonal Fyn-associated substrate |
| chr7_-_126986285 | 0.01 |
ENSMUST00000164345.2
|
9130019O22Rik
|
RIKEN cDNA 9130019O22 gene |
| chr5_-_35836761 | 0.01 |
ENSMUST00000114233.3
|
Htra3
|
HtrA serine peptidase 3 |
| chr10_-_78588884 | 0.01 |
ENSMUST00000105383.4
|
Ccdc105
|
coiled-coil domain containing 105 |
| chr5_-_135280063 | 0.01 |
ENSMUST00000062572.3
|
Fzd9
|
frizzled class receptor 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Xbp1_Creb3l1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 0.2 | 0.6 | GO:0060435 | bronchiole development(GO:0060435) intestinal epithelial cell maturation(GO:0060574) |
| 0.2 | 1.8 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.2 | 0.6 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.2 | 0.5 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.2 | 0.6 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.9 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 0.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.5 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 0.8 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.2 | GO:0002396 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.1 | 0.3 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.1 | 1.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.6 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 2.9 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.1 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.0 | 0.2 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.2 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.0 | 0.3 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.4 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.8 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0002880 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.0 | 0.1 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.0 | 0.3 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:1903297 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.2 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.0 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.7 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 1.0 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 1.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 1.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 1.5 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.2 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 1.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.0 | GO:0097209 | epidermal lamellar body(GO:0097209) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.2 | 0.5 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.1 | 0.8 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 2.9 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.6 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.3 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 4.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.3 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.1 | 0.4 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.8 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 1.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.5 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0038100 | nodal binding(GO:0038100) |
| 0.0 | 0.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.0 | 0.0 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.1 | GO:0015321 | sodium:inorganic phosphate symporter activity(GO:0015319) sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.8 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |