Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
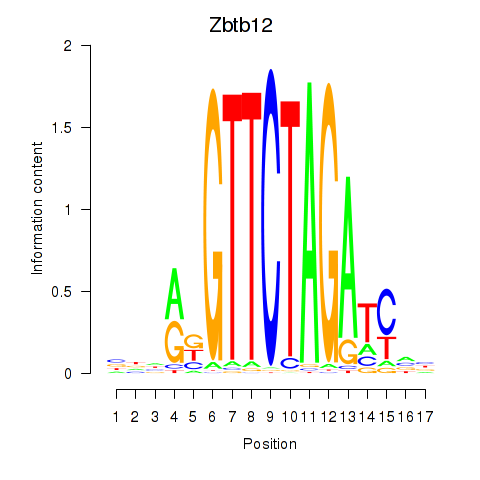
Results for Zbtb12
Z-value: 1.18
Transcription factors associated with Zbtb12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb12
|
ENSMUSG00000049823.10 | zinc finger and BTB domain containing 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb12 | mm39_v1_chr17_+_35113490_35113554 | -0.56 | 3.3e-01 | Click! |
Activity profile of Zbtb12 motif
Sorted Z-values of Zbtb12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_129854256 | 1.09 |
ENSMUST00000110299.3
|
Tgm3
|
transglutaminase 3, E polypeptide |
| chrX_+_93768175 | 0.83 |
ENSMUST00000101388.4
|
Zxdb
|
zinc finger, X-linked, duplicated B |
| chr3_-_131138541 | 0.76 |
ENSMUST00000090246.5
ENSMUST00000126569.2 |
Sgms2
|
sphingomyelin synthase 2 |
| chr11_-_98436626 | 0.60 |
ENSMUST00000103141.4
|
Ikzf3
|
IKAROS family zinc finger 3 |
| chr7_-_25239229 | 0.59 |
ENSMUST00000044547.10
ENSMUST00000066503.14 ENSMUST00000064862.13 |
Ceacam2
|
carcinoembryonic antigen-related cell adhesion molecule 2 |
| chr7_+_99825886 | 0.54 |
ENSMUST00000178946.9
|
Kcne3
|
potassium voltage-gated channel, Isk-related subfamily, gene 3 |
| chr13_-_22219738 | 0.54 |
ENSMUST00000091742.6
|
H2ac12
|
H2A clustered histone 12 |
| chr7_-_25176959 | 0.52 |
ENSMUST00000098668.3
ENSMUST00000206687.2 ENSMUST00000206676.2 ENSMUST00000205308.2 ENSMUST00000098669.8 ENSMUST00000206171.2 ENSMUST00000098666.9 |
Ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr11_+_54793569 | 0.44 |
ENSMUST00000082430.11
|
Gpx3
|
glutathione peroxidase 3 |
| chr13_+_9326513 | 0.39 |
ENSMUST00000174552.8
|
Dip2c
|
disco interacting protein 2 homolog C |
| chr9_+_38407097 | 0.37 |
ENSMUST00000214003.3
ENSMUST00000214264.3 |
Olfr907
|
olfactory receptor 907 |
| chr17_+_48606948 | 0.33 |
ENSMUST00000233092.2
|
Treml2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr14_-_79718890 | 0.27 |
ENSMUST00000022601.7
|
Wbp4
|
WW domain binding protein 4 |
| chr7_-_126574959 | 0.25 |
ENSMUST00000206296.2
ENSMUST00000205437.3 |
Cdiptos
|
CDIP transferase, opposite strand |
| chr11_+_69881885 | 0.24 |
ENSMUST00000018711.15
|
Gabarap
|
gamma-aminobutyric acid receptor associated protein |
| chr8_-_43981143 | 0.22 |
ENSMUST00000080135.5
|
Adam26b
|
a disintegrin and metallopeptidase domain 26B |
| chr6_-_24528012 | 0.22 |
ENSMUST00000023851.9
|
Ndufa5
|
NADH:ubiquinone oxidoreductase subunit A5 |
| chr4_-_41723129 | 0.20 |
ENSMUST00000171641.2
ENSMUST00000030158.11 |
Dctn3
|
dynactin 3 |
| chr15_-_82238432 | 0.20 |
ENSMUST00000230494.2
ENSMUST00000023085.7 |
Ndufa6
|
NADH:ubiquinone oxidoreductase subunit A6 |
| chr5_-_142515792 | 0.20 |
ENSMUST00000099400.3
|
Papolb
|
poly (A) polymerase beta (testis specific) |
| chr14_+_54380308 | 0.17 |
ENSMUST00000196323.2
|
Trdc
|
T cell receptor delta, constant region |
| chr16_-_19703014 | 0.16 |
ENSMUST00000100083.5
ENSMUST00000231564.2 |
A930003A15Rik
|
RIKEN cDNA A930003A15 gene |
| chr9_-_62895197 | 0.16 |
ENSMUST00000216209.2
|
Pias1
|
protein inhibitor of activated STAT 1 |
| chr16_-_20121108 | 0.15 |
ENSMUST00000048642.15
ENSMUST00000232036.2 |
Parl
|
presenilin associated, rhomboid-like |
| chr5_+_96989853 | 0.15 |
ENSMUST00000196126.2
|
Anxa3
|
annexin A3 |
| chr10_-_5144699 | 0.15 |
ENSMUST00000215467.2
|
Syne1
|
spectrin repeat containing, nuclear envelope 1 |
| chr10_+_75152705 | 0.15 |
ENSMUST00000105420.3
|
Adora2a
|
adenosine A2a receptor |
| chr16_+_55786638 | 0.15 |
ENSMUST00000023269.5
|
Rpl24
|
ribosomal protein L24 |
| chr11_-_70590923 | 0.14 |
ENSMUST00000108543.4
ENSMUST00000108542.8 ENSMUST00000108541.9 ENSMUST00000126114.9 ENSMUST00000073625.8 |
Inca1
|
inhibitor of CDK, cyclin A1 interacting protein 1 |
| chr7_+_35285657 | 0.13 |
ENSMUST00000040844.16
ENSMUST00000188906.7 ENSMUST00000186245.7 ENSMUST00000190503.7 |
Ankrd27
|
ankyrin repeat domain 27 (VPS9 domain) |
| chr4_+_43669266 | 0.13 |
ENSMUST00000107864.8
|
Tmem8b
|
transmembrane protein 8B |
| chr9_+_123902143 | 0.12 |
ENSMUST00000168841.3
ENSMUST00000055918.7 |
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr11_+_54793743 | 0.11 |
ENSMUST00000149324.3
|
Gpx3
|
glutathione peroxidase 3 |
| chr7_+_126575510 | 0.11 |
ENSMUST00000206780.2
|
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr19_-_46561532 | 0.11 |
ENSMUST00000026009.10
ENSMUST00000236255.2 |
Arl3
|
ADP-ribosylation factor-like 3 |
| chr14_+_26414422 | 0.11 |
ENSMUST00000022433.12
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr13_-_14787602 | 0.10 |
ENSMUST00000220621.2
|
Mrpl32
|
mitochondrial ribosomal protein L32 |
| chr19_+_6130059 | 0.10 |
ENSMUST00000149347.8
ENSMUST00000143303.2 |
Tmem262
|
transmembrane protein 262 |
| chr8_-_70411054 | 0.10 |
ENSMUST00000211960.2
|
Gatad2a
|
GATA zinc finger domain containing 2A |
| chr18_+_36877709 | 0.10 |
ENSMUST00000007042.6
ENSMUST00000237095.2 |
Ik
|
IK cytokine |
| chr11_-_118021460 | 0.10 |
ENSMUST00000132685.9
|
Dnah17
|
dynein, axonemal, heavy chain 17 |
| chr19_+_6919691 | 0.09 |
ENSMUST00000113426.8
ENSMUST00000238184.2 ENSMUST00000113423.10 ENSMUST00000237334.2 ENSMUST00000235240.2 |
Bad
|
BCL2-associated agonist of cell death |
| chr17_-_15596230 | 0.09 |
ENSMUST00000014917.8
|
Dll1
|
delta like canonical Notch ligand 1 |
| chrX_-_139857424 | 0.09 |
ENSMUST00000033805.15
ENSMUST00000112978.2 |
Psmd10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr14_-_104081827 | 0.08 |
ENSMUST00000022718.11
|
Ednrb
|
endothelin receptor type B |
| chr6_+_108041998 | 0.08 |
ENSMUST00000049246.7
|
Setmar
|
SET domain without mariner transposase fusion |
| chr4_+_119396913 | 0.08 |
ENSMUST00000137560.8
|
Foxj3
|
forkhead box J3 |
| chr12_+_86725459 | 0.08 |
ENSMUST00000021681.4
|
Vash1
|
vasohibin 1 |
| chr11_-_59118988 | 0.07 |
ENSMUST00000163300.8
ENSMUST00000061242.8 |
Arf1
|
ADP-ribosylation factor 1 |
| chr1_+_38037086 | 0.07 |
ENSMUST00000027252.8
|
Eif5b
|
eukaryotic translation initiation factor 5B |
| chr2_-_150531280 | 0.07 |
ENSMUST00000046095.10
|
Vsx1
|
visual system homeobox 1 |
| chr9_+_22322802 | 0.06 |
ENSMUST00000058868.9
|
9530077C05Rik
|
RIKEN cDNA 9530077C05 gene |
| chr18_-_20995980 | 0.05 |
ENSMUST00000224530.2
|
Trappc8
|
trafficking protein particle complex 8 |
| chr15_-_99372575 | 0.05 |
ENSMUST00000040313.6
|
Bcdin3d
|
BCDIN3 domain containing |
| chr5_-_22070602 | 0.05 |
ENSMUST00000030878.8
|
Slc26a5
|
solute carrier family 26, member 5 |
| chr17_+_18051266 | 0.05 |
ENSMUST00000172097.9
|
Spaca6
|
sperm acrosome associated 6 |
| chr11_+_98337655 | 0.05 |
ENSMUST00000019456.5
|
Grb7
|
growth factor receptor bound protein 7 |
| chr9_+_5308828 | 0.05 |
ENSMUST00000162846.8
ENSMUST00000027012.14 |
Casp4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr4_-_43669141 | 0.04 |
ENSMUST00000056474.7
|
Fam221b
|
family with sequence similarity 221, member B |
| chr17_-_24866749 | 0.04 |
ENSMUST00000234121.2
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr11_-_50806962 | 0.04 |
ENSMUST00000116378.8
ENSMUST00000109128.8 |
Zfp2
|
zinc finger protein 2 |
| chr13_-_64645606 | 0.04 |
ENSMUST00000180282.2
|
Fam240b
|
family with sequence similarity 240 member B |
| chr4_-_149994176 | 0.04 |
ENSMUST00000105685.2
|
Spsb1
|
splA/ryanodine receptor domain and SOCS box containing 1 |
| chr11_+_102775991 | 0.04 |
ENSMUST00000100369.4
|
Fam187a
|
family with sequence similarity 187, member A |
| chr1_+_69866145 | 0.04 |
ENSMUST00000065425.12
ENSMUST00000113940.4 |
Spag16
|
sperm associated antigen 16 |
| chr14_+_8555284 | 0.04 |
ENSMUST00000144914.3
|
Gm281
|
predicted gene 281 |
| chr11_+_49138278 | 0.04 |
ENSMUST00000109194.2
|
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chr16_-_94657531 | 0.04 |
ENSMUST00000232562.2
ENSMUST00000165538.3 |
Kcnj6
|
potassium inwardly-rectifying channel, subfamily J, member 6 |
| chr15_+_82225380 | 0.04 |
ENSMUST00000050349.3
|
Pheta2
|
PH domain containing endocytic trafficking adaptor 2 |
| chr17_+_37253802 | 0.04 |
ENSMUST00000040498.12
|
Rnf39
|
ring finger protein 39 |
| chr18_+_70605691 | 0.04 |
ENSMUST00000164223.8
|
Stard6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr15_-_76259126 | 0.03 |
ENSMUST00000071119.8
|
Tssk5
|
testis-specific serine kinase 5 |
| chr8_-_44029744 | 0.03 |
ENSMUST00000049577.3
|
Adam26a
|
a disintegrin and metallopeptidase domain 26A (testase 3) |
| chr6_+_29398919 | 0.03 |
ENSMUST00000181464.8
ENSMUST00000180829.8 |
Ccdc136
|
coiled-coil domain containing 136 |
| chr9_-_44891626 | 0.03 |
ENSMUST00000002101.12
ENSMUST00000160886.2 |
Cd3g
|
CD3 antigen, gamma polypeptide |
| chr15_-_60793115 | 0.03 |
ENSMUST00000096418.5
|
A1bg
|
alpha-1-B glycoprotein |
| chr18_+_70605722 | 0.03 |
ENSMUST00000174118.8
|
Stard6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr16_-_33787399 | 0.03 |
ENSMUST00000023510.7
|
Umps
|
uridine monophosphate synthetase |
| chr9_+_36604516 | 0.03 |
ENSMUST00000034620.5
|
Acrv1
|
acrosomal vesicle protein 1 |
| chr1_-_161807205 | 0.03 |
ENSMUST00000162676.2
|
4930558K02Rik
|
RIKEN cDNA 4930558K02 gene |
| chr6_+_24528143 | 0.03 |
ENSMUST00000031696.10
|
Asb15
|
ankyrin repeat and SOCS box-containing 15 |
| chr13_-_104365310 | 0.03 |
ENSMUST00000022226.6
|
Ppwd1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr14_-_52728503 | 0.03 |
ENSMUST00000073571.6
|
Olfr1507
|
olfactory receptor 1507 |
| chr15_-_101694895 | 0.03 |
ENSMUST00000071104.6
|
Krt72
|
keratin 72 |
| chr14_-_70449438 | 0.03 |
ENSMUST00000227929.2
|
Sorbs3
|
sorbin and SH3 domain containing 3 |
| chr7_+_140427711 | 0.02 |
ENSMUST00000026555.12
|
Odf3
|
outer dense fiber of sperm tails 3 |
| chr7_+_140427729 | 0.02 |
ENSMUST00000106049.2
|
Odf3
|
outer dense fiber of sperm tails 3 |
| chr17_+_48607405 | 0.02 |
ENSMUST00000170941.3
|
Treml2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr8_+_71156071 | 0.02 |
ENSMUST00000212436.2
|
Iqcn
|
IQ motif containing N |
| chr15_-_88838605 | 0.02 |
ENSMUST00000109371.8
|
Ttll8
|
tubulin tyrosine ligase-like family, member 8 |
| chr10_+_62896492 | 0.02 |
ENSMUST00000219687.2
ENSMUST00000219045.2 |
Pbld1
|
phenazine biosynthesis-like protein domain containing 1 |
| chr19_-_12069216 | 0.02 |
ENSMUST00000208703.3
ENSMUST00000219005.2 ENSMUST00000219155.2 ENSMUST00000217952.2 ENSMUST00000112952.4 |
Olfr1426
|
olfactory receptor 1426 |
| chr9_+_37278647 | 0.02 |
ENSMUST00000051839.9
|
Hepacam
|
hepatocyte cell adhesion molecule |
| chr8_+_41246310 | 0.02 |
ENSMUST00000056331.8
|
Adam20
|
a disintegrin and metallopeptidase domain 20 |
| chr19_-_57170725 | 0.02 |
ENSMUST00000133369.2
|
Ablim1
|
actin-binding LIM protein 1 |
| chr7_+_102420428 | 0.02 |
ENSMUST00000213432.2
|
Olfr561
|
olfactory receptor 561 |
| chr9_-_40366966 | 0.02 |
ENSMUST00000165104.8
ENSMUST00000045682.7 |
Gramd1b
|
GRAM domain containing 1B |
| chr5_+_30486375 | 0.02 |
ENSMUST00000101448.5
|
Drc1
|
dynein regulatory complex subunit 1 |
| chr14_+_26789345 | 0.02 |
ENSMUST00000226105.2
|
Il17rd
|
interleukin 17 receptor D |
| chr9_-_58448224 | 0.02 |
ENSMUST00000039788.11
|
Cd276
|
CD276 antigen |
| chr8_+_41205245 | 0.02 |
ENSMUST00000096663.5
|
Adam25
|
a disintegrin and metallopeptidase domain 25 (testase 2) |
| chr17_+_37253916 | 0.02 |
ENSMUST00000173072.2
|
Rnf39
|
ring finger protein 39 |
| chr7_-_141472218 | 0.02 |
ENSMUST00000151890.3
|
Tollip
|
toll interacting protein |
| chr6_+_90439544 | 0.02 |
ENSMUST00000032174.12
|
Klf15
|
Kruppel-like factor 15 |
| chr6_+_90439596 | 0.02 |
ENSMUST00000203039.3
|
Klf15
|
Kruppel-like factor 15 |
| chr19_+_13742643 | 0.02 |
ENSMUST00000215930.2
|
Olfr1495
|
olfactory receptor 1495 |
| chr13_-_92620507 | 0.02 |
ENSMUST00000040106.9
|
Fam151b
|
family with sequence similarity 151, member B |
| chr10_-_70428611 | 0.02 |
ENSMUST00000162251.8
|
Phyhipl
|
phytanoyl-CoA hydroxylase interacting protein-like |
| chr7_-_111758241 | 0.02 |
ENSMUST00000033036.7
|
Dkk3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr5_-_35836761 | 0.02 |
ENSMUST00000114233.3
|
Htra3
|
HtrA serine peptidase 3 |
| chrX_+_76908364 | 0.02 |
ENSMUST00000114039.3
|
Gm14744
|
predicted gene 14744 |
| chr18_+_70605630 | 0.02 |
ENSMUST00000168249.9
|
Stard6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr9_+_39642274 | 0.02 |
ENSMUST00000217257.2
|
Olfr150
|
olfactory receptor 150 |
| chr8_+_41276027 | 0.01 |
ENSMUST00000066814.7
|
Adam39
|
a disintegrin and metallopeptidase domain 39 |
| chr3_+_5283606 | 0.01 |
ENSMUST00000026284.13
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr7_-_48493388 | 0.01 |
ENSMUST00000167786.4
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chrX_-_76968668 | 0.01 |
ENSMUST00000063127.4
|
5430402E10Rik
|
RIKEN cDNA 5430402E10 gene |
| chr9_+_110673565 | 0.01 |
ENSMUST00000176403.8
|
Prss46
|
protease, serine 46 |
| chr7_+_106413336 | 0.01 |
ENSMUST00000166880.3
ENSMUST00000075414.8 |
Olfr701
|
olfactory receptor 701 |
| chr19_-_12069012 | 0.01 |
ENSMUST00000220005.2
|
Olfr1426
|
olfactory receptor 1426 |
| chr5_+_11234174 | 0.01 |
ENSMUST00000168407.3
|
Gm5861
|
predicted gene 5861 |
| chr11_-_100332619 | 0.01 |
ENSMUST00000107398.8
|
Nt5c3b
|
5'-nucleotidase, cytosolic IIIB |
| chrX_+_106836189 | 0.01 |
ENSMUST00000101292.9
|
Tent5d
|
terminal nucleotidyltransferase 5D |
| chr19_-_8196196 | 0.01 |
ENSMUST00000113298.9
|
Slc22a29
|
solute carrier family 22. member 29 |
| chr11_+_100332709 | 0.01 |
ENSMUST00000001599.4
ENSMUST00000107395.3 |
Klhl10
|
kelch-like 10 |
| chr11_+_104983022 | 0.01 |
ENSMUST00000021029.6
|
Efcab3
|
EF-hand calcium binding domain 3 |
| chr12_+_111540920 | 0.01 |
ENSMUST00000075281.8
ENSMUST00000084953.13 |
Mark3
|
MAP/microtubule affinity regulating kinase 3 |
| chr6_-_83504756 | 0.01 |
ENSMUST00000152029.2
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr8_-_107007596 | 0.01 |
ENSMUST00000212896.2
|
Smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr6_-_143892814 | 0.01 |
ENSMUST00000124233.7
ENSMUST00000144289.4 ENSMUST00000111748.8 |
Sox5
|
SRY (sex determining region Y)-box 5 |
| chr3_+_5283577 | 0.01 |
ENSMUST00000175866.8
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr3_-_153610368 | 0.01 |
ENSMUST00000188338.7
|
Msh4
|
mutS homolog 4 |
| chr15_-_76419131 | 0.01 |
ENSMUST00000230604.2
|
Tmem249
|
transmembrane protein 249 |
| chr10_+_63293284 | 0.01 |
ENSMUST00000105440.8
|
Ctnna3
|
catenin (cadherin associated protein), alpha 3 |
| chr6_+_103674695 | 0.01 |
ENSMUST00000205098.2
|
Chl1
|
cell adhesion molecule L1-like |
| chr3_-_153610528 | 0.01 |
ENSMUST00000190449.2
|
Msh4
|
mutS homolog 4 |
| chr6_-_68784692 | 0.00 |
ENSMUST00000103334.4
|
Igkv4-90
|
immunoglobulin kappa chain variable 4-90 |
| chr17_-_28128180 | 0.00 |
ENSMUST00000114848.8
|
Taf11
|
TATA-box binding protein associated factor 11 |
| chr1_-_75208734 | 0.00 |
ENSMUST00000180101.3
|
A630095N17Rik
|
RIKEN cDNA A630095N17 gene |
| chr19_+_12186932 | 0.00 |
ENSMUST00000072316.5
|
Olfr1431
|
olfactory receptor 1431 |
| chr8_+_41128099 | 0.00 |
ENSMUST00000051614.5
|
Adam24
|
a disintegrin and metallopeptidase domain 24 (testase 1) |
| chr4_-_138351313 | 0.00 |
ENSMUST00000030533.12
|
Vwa5b1
|
von Willebrand factor A domain containing 5B1 |
| chr1_-_155627430 | 0.00 |
ENSMUST00000195275.2
|
Lhx4
|
LIM homeobox protein 4 |
| chr18_+_70605476 | 0.00 |
ENSMUST00000114959.9
|
Stard6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr5_-_121641461 | 0.00 |
ENSMUST00000079368.5
|
Adam1b
|
a disintegrin and metallopeptidase domain 1b |
| chr7_-_48145873 | 0.00 |
ENSMUST00000186394.2
|
Mrgprx2
|
MAS-related GPR, member X2 |
| chr11_+_49426014 | 0.00 |
ENSMUST00000074543.2
|
Olfr1382
|
olfactory receptor 1382 |
| chr17_+_27248233 | 0.00 |
ENSMUST00000053683.7
ENSMUST00000236222.2 |
Ggnbp1
|
gametogenetin binding protein 1 |
| chr10_-_75658355 | 0.00 |
ENSMUST00000160211.2
|
Gstt4
|
glutathione S-transferase, theta 4 |
| chr7_+_65693350 | 0.00 |
ENSMUST00000206065.2
|
Gm45213
|
predicted gene 45213 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb12
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0070237 | positive regulation of activation-induced cell death of T cells(GO:0070237) |
| 0.1 | 0.5 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.8 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.6 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.1 | GO:0002436 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) negative regulation of eosinophil activation(GO:1902567) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) |
| 0.0 | 1.1 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.1 | GO:1902220 | positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902220) |
| 0.0 | 0.1 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.0 | 0.1 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0097212 | cleavage furrow ingression(GO:0036090) lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.1 | GO:0015755 | fructose transport(GO:0015755) |
| 0.0 | 0.6 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.0 | 0.0 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 1.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.0 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 1.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.0 | 0.6 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.0 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |