Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Zbtb16
Z-value: 1.11
Transcription factors associated with Zbtb16
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb16
|
ENSMUSG00000066687.6 | zinc finger and BTB domain containing 16 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb16 | mm39_v1_chr9_-_48747232_48747262 | -0.65 | 2.3e-01 | Click! |
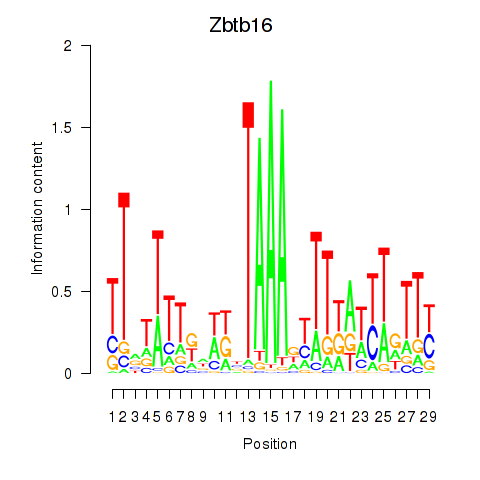
Activity profile of Zbtb16 motif
Sorted Z-values of Zbtb16 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_75678092 | 1.45 |
ENSMUST00000238938.2
|
Rplp2-ps1
|
ribosomal protein, large P2, pseudogene 1 |
| chr15_+_79555272 | 0.86 |
ENSMUST00000127292.2
|
Tomm22
|
translocase of outer mitochondrial membrane 22 |
| chr11_+_23615612 | 0.75 |
ENSMUST00000109525.8
ENSMUST00000020520.11 |
Pus10
|
pseudouridylate synthase 10 |
| chr11_+_87685032 | 0.68 |
ENSMUST00000121303.8
|
Mpo
|
myeloperoxidase |
| chr11_-_93856783 | 0.67 |
ENSMUST00000021220.10
|
Nme1
|
NME/NM23 nucleoside diphosphate kinase 1 |
| chr4_-_117035922 | 0.63 |
ENSMUST00000153953.2
ENSMUST00000106436.8 |
Kif2c
|
kinesin family member 2C |
| chr5_-_121045568 | 0.63 |
ENSMUST00000080322.8
|
Oas1a
|
2'-5' oligoadenylate synthetase 1A |
| chr13_+_54225828 | 0.63 |
ENSMUST00000021930.10
|
Sfxn1
|
sideroflexin 1 |
| chr4_+_116544509 | 0.63 |
ENSMUST00000030454.6
|
Prdx1
|
peroxiredoxin 1 |
| chr6_+_40619913 | 0.62 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chr3_+_32760447 | 0.62 |
ENSMUST00000194781.6
|
Actl6a
|
actin-like 6A |
| chr17_-_27816151 | 0.56 |
ENSMUST00000231742.2
|
Nudt3
|
nudix (nucleotide diphosphate linked moiety X)-type motif 3 |
| chr5_+_138159333 | 0.52 |
ENSMUST00000019638.15
ENSMUST00000110951.8 |
Cops6
|
COP9 signalosome subunit 6 |
| chr4_-_107975701 | 0.50 |
ENSMUST00000149106.8
|
Scp2
|
sterol carrier protein 2, liver |
| chr6_-_30390996 | 0.49 |
ENSMUST00000152391.9
ENSMUST00000115184.2 ENSMUST00000080812.14 ENSMUST00000102992.10 |
Zc3hc1
|
zinc finger, C3HC type 1 |
| chr2_+_125994050 | 0.46 |
ENSMUST00000170908.8
|
Dtwd1
|
DTW domain containing 1 |
| chr3_-_146475974 | 0.46 |
ENSMUST00000106137.8
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr4_-_107975723 | 0.45 |
ENSMUST00000030340.15
|
Scp2
|
sterol carrier protein 2, liver |
| chr6_+_137731526 | 0.44 |
ENSMUST00000203216.3
ENSMUST00000087675.9 ENSMUST00000203693.3 |
Dera
|
deoxyribose-phosphate aldolase (putative) |
| chr17_+_46807637 | 0.44 |
ENSMUST00000046497.8
|
Dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr17_-_36343573 | 0.43 |
ENSMUST00000102678.5
|
H2-T23
|
histocompatibility 2, T region locus 23 |
| chr19_-_32717166 | 0.42 |
ENSMUST00000235142.2
ENSMUST00000070210.6 ENSMUST00000236011.2 |
Atad1
|
ATPase family, AAA domain containing 1 |
| chr9_+_19828161 | 0.41 |
ENSMUST00000217347.2
ENSMUST00000057596.10 |
Olfr77
|
olfactory receptor 77 |
| chr5_+_110478558 | 0.40 |
ENSMUST00000112481.2
|
Pole
|
polymerase (DNA directed), epsilon |
| chr17_+_47680097 | 0.40 |
ENSMUST00000060752.13
ENSMUST00000119841.8 |
Mrps10
|
mitochondrial ribosomal protein S10 |
| chr11_-_48717482 | 0.39 |
ENSMUST00000104959.2
|
Gm12184
|
predicted gene 12184 |
| chr19_-_8691797 | 0.39 |
ENSMUST00000206797.2
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr19_-_32717138 | 0.38 |
ENSMUST00000236985.2
|
Atad1
|
ATPase family, AAA domain containing 1 |
| chr9_+_56844746 | 0.37 |
ENSMUST00000034827.10
|
Imp3
|
IMP3, U3 small nucleolar ribonucleoprotein |
| chr10_-_80736579 | 0.37 |
ENSMUST00000218481.2
ENSMUST00000219896.2 ENSMUST00000020440.7 |
Timm13
|
translocase of inner mitochondrial membrane 13 |
| chr17_-_65901946 | 0.36 |
ENSMUST00000232686.2
|
Vapa
|
vesicle-associated membrane protein, associated protein A |
| chr3_-_105940130 | 0.36 |
ENSMUST00000200146.2
|
Chil5
|
chitinase-like 5 |
| chr4_+_44300876 | 0.36 |
ENSMUST00000045607.12
|
Melk
|
maternal embryonic leucine zipper kinase |
| chr2_-_50186690 | 0.35 |
ENSMUST00000144143.8
ENSMUST00000102769.11 ENSMUST00000133768.2 |
Mmadhc
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria |
| chr7_-_125090540 | 0.35 |
ENSMUST00000138616.3
|
Nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr1_-_149836974 | 0.34 |
ENSMUST00000190507.2
ENSMUST00000070200.15 |
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr19_-_8691460 | 0.34 |
ENSMUST00000206560.2
ENSMUST00000205538.2 |
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr5_+_8848138 | 0.33 |
ENSMUST00000009058.10
|
Abcb1b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1B |
| chr17_+_34131696 | 0.33 |
ENSMUST00000174146.3
|
Daxx
|
Fas death domain-associated protein |
| chr16_+_95946591 | 0.32 |
ENSMUST00000023913.11
ENSMUST00000232832.2 ENSMUST00000233566.2 ENSMUST00000233273.2 |
Get1
|
guided entry of tail-anchored proteins factor 1 |
| chr6_+_134617903 | 0.32 |
ENSMUST00000062755.10
|
Borcs5
|
BLOC-1 related complex subunit 5 |
| chr10_-_93727003 | 0.32 |
ENSMUST00000180840.8
|
Metap2
|
methionine aminopeptidase 2 |
| chr9_-_122695071 | 0.32 |
ENSMUST00000216063.2
|
Zfp445
|
zinc finger protein 445 |
| chr8_+_27532623 | 0.32 |
ENSMUST00000209856.2
ENSMUST00000098851.12 ENSMUST00000211393.2 ENSMUST00000211518.2 |
Plpbp
|
pyridoxal phosphate binding protein |
| chr4_+_123176570 | 0.32 |
ENSMUST00000106243.8
ENSMUST00000106241.8 ENSMUST00000080178.13 |
Pabpc4
|
poly(A) binding protein, cytoplasmic 4 |
| chr17_-_71305003 | 0.31 |
ENSMUST00000024846.13
ENSMUST00000232766.2 |
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr18_-_67582191 | 0.31 |
ENSMUST00000025408.10
|
Afg3l2
|
AFG3-like AAA ATPase 2 |
| chr18_-_35795233 | 0.31 |
ENSMUST00000025209.12
ENSMUST00000096573.4 |
Spata24
|
spermatogenesis associated 24 |
| chr7_-_16121682 | 0.31 |
ENSMUST00000094815.5
|
Sae1
|
SUMO1 activating enzyme subunit 1 |
| chr9_-_118986123 | 0.31 |
ENSMUST00000010795.5
|
Acaa1b
|
acetyl-Coenzyme A acyltransferase 1B |
| chr11_-_106889291 | 0.30 |
ENSMUST00000124541.8
|
Kpna2
|
karyopherin (importin) alpha 2 |
| chr11_+_68936457 | 0.30 |
ENSMUST00000108666.8
ENSMUST00000021277.6 |
Aurkb
|
aurora kinase B |
| chr8_+_70625032 | 0.29 |
ENSMUST00000002413.15
ENSMUST00000182980.8 ENSMUST00000182365.8 |
Tmem161a
|
transmembrane protein 161A |
| chr14_-_73785682 | 0.29 |
ENSMUST00000043813.3
|
Nudt15
|
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr13_-_8921732 | 0.28 |
ENSMUST00000054251.13
ENSMUST00000176813.8 ENSMUST00000175958.2 |
Wdr37
|
WD repeat domain 37 |
| chr6_-_129599645 | 0.28 |
ENSMUST00000032252.8
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr19_-_8763771 | 0.28 |
ENSMUST00000176496.8
|
Taf6l
|
TATA-box binding protein associated factor 6 like |
| chr5_+_115061293 | 0.28 |
ENSMUST00000031540.11
ENSMUST00000112143.4 |
Oasl1
|
2'-5' oligoadenylate synthetase-like 1 |
| chr7_-_133384449 | 0.27 |
ENSMUST00000063669.8
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr1_-_93659622 | 0.27 |
ENSMUST00000189728.7
|
Thap4
|
THAP domain containing 4 |
| chr11_-_115590318 | 0.27 |
ENSMUST00000106497.8
|
Grb2
|
growth factor receptor bound protein 2 |
| chr17_+_26471870 | 0.27 |
ENSMUST00000025023.15
|
Luc7l
|
Luc7-like |
| chr9_+_62754252 | 0.27 |
ENSMUST00000124984.2
|
Cln6
|
ceroid-lipofuscinosis, neuronal 6 |
| chr17_+_29493113 | 0.27 |
ENSMUST00000234326.2
ENSMUST00000235117.2 |
BC004004
|
cDNA sequence BC004004 |
| chr2_-_32321116 | 0.26 |
ENSMUST00000127961.3
ENSMUST00000136361.8 ENSMUST00000052119.14 |
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr2_-_86180622 | 0.26 |
ENSMUST00000099894.5
ENSMUST00000213564.3 |
Olfr1055
|
olfactory receptor 1055 |
| chr1_-_23436620 | 0.26 |
ENSMUST00000188677.2
|
Ogfrl1
|
opioid growth factor receptor-like 1 |
| chr1_+_165616250 | 0.26 |
ENSMUST00000161971.8
ENSMUST00000187313.7 ENSMUST00000178336.8 ENSMUST00000005907.12 ENSMUST00000027849.11 |
Cd247
|
CD247 antigen |
| chr7_-_125090757 | 0.26 |
ENSMUST00000033006.14
|
Nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr12_+_55286111 | 0.25 |
ENSMUST00000164243.2
|
Srp54c
|
signal recognition particle 54C |
| chr2_-_25255650 | 0.25 |
ENSMUST00000102925.4
|
Uap1l1
|
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1 |
| chr7_-_16121716 | 0.25 |
ENSMUST00000211741.2
ENSMUST00000210999.2 |
Sae1
|
SUMO1 activating enzyme subunit 1 |
| chr8_+_85635189 | 0.25 |
ENSMUST00000003910.13
ENSMUST00000109744.8 |
Dnase2a
|
deoxyribonuclease II alpha |
| chr9_+_104930438 | 0.25 |
ENSMUST00000149243.8
ENSMUST00000035177.15 ENSMUST00000214036.2 |
Mrpl3
|
mitochondrial ribosomal protein L3 |
| chr5_-_23821523 | 0.24 |
ENSMUST00000088392.9
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr6_+_29272625 | 0.24 |
ENSMUST00000054445.9
|
Hilpda
|
hypoxia inducible lipid droplet associated |
| chr7_+_101520843 | 0.24 |
ENSMUST00000210984.2
|
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr17_+_26471889 | 0.24 |
ENSMUST00000114976.9
ENSMUST00000140427.8 ENSMUST00000119928.8 |
Luc7l
|
Luc7-like |
| chr13_+_30320446 | 0.24 |
ENSMUST00000047311.16
|
Mboat1
|
membrane bound O-acyltransferase domain containing 1 |
| chr11_-_50093781 | 0.24 |
ENSMUST00000136936.2
|
Sqstm1
|
sequestosome 1 |
| chr4_-_119177614 | 0.24 |
ENSMUST00000147077.8
ENSMUST00000056458.14 ENSMUST00000106321.9 ENSMUST00000106319.8 ENSMUST00000106317.2 ENSMUST00000106318.8 |
Ppih
|
peptidyl prolyl isomerase H |
| chr6_-_129655868 | 0.24 |
ENSMUST00000118447.2
ENSMUST00000169545.8 ENSMUST00000032270.13 ENSMUST00000032271.13 |
Klrc1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr1_+_157334298 | 0.23 |
ENSMUST00000086130.9
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr18_-_35795175 | 0.23 |
ENSMUST00000236574.2
ENSMUST00000236971.2 |
Spata24
|
spermatogenesis associated 24 |
| chr1_+_157334347 | 0.23 |
ENSMUST00000027881.15
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chrY_-_1286623 | 0.23 |
ENSMUST00000091190.12
|
Ddx3y
|
DEAD box helicase 3, Y-linked |
| chr17_+_29493049 | 0.23 |
ENSMUST00000149405.4
|
BC004004
|
cDNA sequence BC004004 |
| chr11_-_17903861 | 0.23 |
ENSMUST00000076661.7
|
Etaa1
|
Ewing tumor-associated antigen 1 |
| chr11_-_101308441 | 0.22 |
ENSMUST00000070395.9
|
Aarsd1
|
alanyl-tRNA synthetase domain containing 1 |
| chr7_+_27259895 | 0.22 |
ENSMUST00000187032.2
|
2310022A10Rik
|
RIKEN cDNA 2310022A10 gene |
| chr16_-_56533179 | 0.22 |
ENSMUST00000136394.8
|
Tfg
|
Trk-fused gene |
| chr8_+_27532583 | 0.22 |
ENSMUST00000033875.10
ENSMUST00000209525.2 |
Plpbp
|
pyridoxal phosphate binding protein |
| chr11_-_83469446 | 0.22 |
ENSMUST00000019266.6
|
Ccl9
|
chemokine (C-C motif) ligand 9 |
| chr5_-_77053310 | 0.22 |
ENSMUST00000146570.8
ENSMUST00000142450.2 ENSMUST00000120963.8 |
Aasdh
|
aminoadipate-semialdehyde dehydrogenase |
| chr14_-_66071412 | 0.21 |
ENSMUST00000022613.10
|
Esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr4_-_126094910 | 0.21 |
ENSMUST00000136157.8
|
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr15_-_96597610 | 0.21 |
ENSMUST00000023099.8
|
Slc38a2
|
solute carrier family 38, member 2 |
| chr3_+_89110223 | 0.21 |
ENSMUST00000077367.11
ENSMUST00000167998.2 |
Gba
|
glucosidase, beta, acid |
| chr2_-_15054065 | 0.21 |
ENSMUST00000028034.15
ENSMUST00000114713.2 |
Nsun6
|
NOL1/NOP2/Sun domain family member 6 |
| chrX_-_133012600 | 0.21 |
ENSMUST00000033610.13
|
Nox1
|
NADPH oxidase 1 |
| chr5_+_143534455 | 0.21 |
ENSMUST00000169329.8
ENSMUST00000067145.12 ENSMUST00000119488.2 ENSMUST00000118121.2 ENSMUST00000200267.2 ENSMUST00000196487.2 |
Fam220a
Fam220a
|
family with sequence similarity 220, member A family with sequence similarity 220, member A |
| chr17_+_46421908 | 0.20 |
ENSMUST00000024763.10
ENSMUST00000123646.2 |
Mrps18a
|
mitochondrial ribosomal protein S18A |
| chr7_+_27869192 | 0.20 |
ENSMUST00000208967.2
|
Fbl
|
fibrillarin |
| chr2_+_30171055 | 0.20 |
ENSMUST00000143119.3
|
Gm28038
|
predicted gene, 28038 |
| chr11_+_65698001 | 0.20 |
ENSMUST00000071465.9
ENSMUST00000018491.8 |
Zkscan6
|
zinc finger with KRAB and SCAN domains 6 |
| chr13_+_55875158 | 0.20 |
ENSMUST00000021958.6
ENSMUST00000124968.8 |
Pcbd2
|
pterin 4 alpha carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2 |
| chr19_-_5507532 | 0.20 |
ENSMUST00000236881.2
|
Ccdc85b
|
coiled-coil domain containing 85B |
| chr4_-_58912678 | 0.20 |
ENSMUST00000144512.8
ENSMUST00000102889.10 ENSMUST00000055822.15 |
Ecpas
|
Ecm29 proteasome adaptor and scaffold |
| chr11_+_101518768 | 0.20 |
ENSMUST00000010506.10
|
Rdm1
|
RAD52 motif 1 |
| chr2_-_120183575 | 0.19 |
ENSMUST00000028752.8
ENSMUST00000102501.10 |
Vps39
|
VPS39 HOPS complex subunit |
| chr4_-_135780660 | 0.19 |
ENSMUST00000102536.11
|
Rpl11
|
ribosomal protein L11 |
| chr4_+_155666933 | 0.19 |
ENSMUST00000105612.2
|
Nadk
|
NAD kinase |
| chr2_+_80469142 | 0.19 |
ENSMUST00000028382.13
ENSMUST00000124377.2 |
Nup35
|
nucleoporin 35 |
| chr3_+_84081411 | 0.18 |
ENSMUST00000193882.2
|
Gm6525
|
predicted pseudogene 6525 |
| chr14_-_55880980 | 0.18 |
ENSMUST00000132338.8
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr8_+_71068791 | 0.18 |
ENSMUST00000210609.2
|
Lrrc25
|
leucine rich repeat containing 25 |
| chr11_-_68899248 | 0.18 |
ENSMUST00000021282.12
|
Pfas
|
phosphoribosylformylglycinamidine synthase (FGAR amidotransferase) |
| chr7_+_27869115 | 0.17 |
ENSMUST00000042405.8
|
Fbl
|
fibrillarin |
| chr1_-_120197979 | 0.17 |
ENSMUST00000112639.8
|
Steap3
|
STEAP family member 3 |
| chr9_+_96141299 | 0.16 |
ENSMUST00000179065.8
|
Tfdp2
|
transcription factor Dp 2 |
| chr5_-_130284366 | 0.16 |
ENSMUST00000026387.11
|
Sbds
|
SBDS ribosome maturation factor |
| chr4_+_129181407 | 0.16 |
ENSMUST00000102599.4
|
Sync
|
syncoilin |
| chr1_+_179936757 | 0.16 |
ENSMUST00000143176.8
ENSMUST00000135056.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr3_-_129518723 | 0.16 |
ENSMUST00000199615.5
ENSMUST00000197079.5 |
Egf
|
epidermal growth factor |
| chr16_-_87229485 | 0.16 |
ENSMUST00000039449.9
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chr17_+_29493157 | 0.16 |
ENSMUST00000234234.2
|
BC004004
|
cDNA sequence BC004004 |
| chr9_+_96141317 | 0.16 |
ENSMUST00000165768.4
|
Tfdp2
|
transcription factor Dp 2 |
| chr9_-_44679136 | 0.15 |
ENSMUST00000034607.10
|
Arcn1
|
archain 1 |
| chr7_-_126611037 | 0.15 |
ENSMUST00000133172.2
|
Mvp
|
major vault protein |
| chr17_+_36190662 | 0.15 |
ENSMUST00000025292.15
|
Dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr7_-_104050027 | 0.15 |
ENSMUST00000106828.3
|
Trim30c
|
tripartite motif-containing 30C |
| chr14_-_55881177 | 0.15 |
ENSMUST00000138085.2
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chrX_+_73352694 | 0.15 |
ENSMUST00000130581.2
|
Gdi1
|
guanosine diphosphate (GDP) dissociation inhibitor 1 |
| chr7_-_127805518 | 0.15 |
ENSMUST00000033049.9
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr19_-_33728759 | 0.15 |
ENSMUST00000147153.4
|
Lipo2
|
lipase, member O2 |
| chr18_+_37858753 | 0.14 |
ENSMUST00000066149.9
|
Pcdhga8
|
protocadherin gamma subfamily A, 8 |
| chr15_-_57982705 | 0.14 |
ENSMUST00000228783.2
|
Atad2
|
ATPase family, AAA domain containing 2 |
| chr15_+_6451721 | 0.14 |
ENSMUST00000163082.2
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr13_+_67052978 | 0.14 |
ENSMUST00000168767.9
|
Gm10767
|
predicted gene 10767 |
| chr17_+_35308206 | 0.14 |
ENSMUST00000007251.14
|
Abhd16a
|
abhydrolase domain containing 16A |
| chr9_+_54493618 | 0.14 |
ENSMUST00000217484.2
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr6_+_94477294 | 0.14 |
ENSMUST00000061118.11
|
Slc25a26
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 26 |
| chr9_+_20799471 | 0.14 |
ENSMUST00000004203.6
|
Ppan
|
peter pan homolog |
| chr7_+_18737944 | 0.13 |
ENSMUST00000053713.5
|
Irf2bp1
|
interferon regulatory factor 2 binding protein 1 |
| chr9_-_79885063 | 0.13 |
ENSMUST00000093811.11
|
Filip1
|
filamin A interacting protein 1 |
| chr6_+_85888850 | 0.13 |
ENSMUST00000200680.4
|
Tprkb
|
Tp53rk binding protein |
| chr18_+_77861656 | 0.13 |
ENSMUST00000114748.2
|
Atp5a1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 |
| chr6_+_29272463 | 0.13 |
ENSMUST00000115289.2
|
Hilpda
|
hypoxia inducible lipid droplet associated |
| chr10_-_116385007 | 0.12 |
ENSMUST00000164088.8
|
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chrX_-_162859429 | 0.12 |
ENSMUST00000134272.2
|
Siah1b
|
siah E3 ubiquitin protein ligase 1B |
| chr1_+_58841808 | 0.12 |
ENSMUST00000190213.2
|
Casp8
|
caspase 8 |
| chr4_+_32623985 | 0.11 |
ENSMUST00000108178.2
|
Casp8ap2
|
caspase 8 associated protein 2 |
| chr19_+_53298906 | 0.11 |
ENSMUST00000003870.15
|
Mxi1
|
MAX interactor 1, dimerization protein |
| chr6_-_114898739 | 0.11 |
ENSMUST00000032459.14
|
Vgll4
|
vestigial like family member 4 |
| chr9_+_96140781 | 0.11 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr6_-_48685108 | 0.11 |
ENSMUST00000126422.3
ENSMUST00000119315.2 ENSMUST00000053661.7 |
Gimap6
|
GTPase, IMAP family member 6 |
| chr4_+_155915729 | 0.10 |
ENSMUST00000139651.8
ENSMUST00000084097.12 |
Aurkaip1
|
aurora kinase A interacting protein 1 |
| chr9_-_122694999 | 0.10 |
ENSMUST00000214558.2
ENSMUST00000216721.2 |
Zfp445
|
zinc finger protein 445 |
| chr12_-_102390000 | 0.10 |
ENSMUST00000110020.8
|
Lgmn
|
legumain |
| chr5_-_104169785 | 0.10 |
ENSMUST00000031251.16
|
Hsd17b11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr12_+_65272287 | 0.10 |
ENSMUST00000046331.5
ENSMUST00000221658.2 |
Wdr20rt
|
WD repeat domain 20, retrogene |
| chr17_-_13213054 | 0.10 |
ENSMUST00000233867.2
|
Wtap
|
Wilms tumour 1-associating protein |
| chr1_+_165616315 | 0.10 |
ENSMUST00000161559.3
|
Cd247
|
CD247 antigen |
| chr14_-_55881257 | 0.09 |
ENSMUST00000122358.8
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr2_-_145776934 | 0.09 |
ENSMUST00000001818.5
|
Crnkl1
|
crooked neck pre-mRNA splicing factor 1 |
| chr7_-_45173193 | 0.09 |
ENSMUST00000211212.2
|
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr18_+_56695515 | 0.09 |
ENSMUST00000130163.8
ENSMUST00000132628.8 |
Phax
|
phosphorylated adaptor for RNA export |
| chr16_-_94172691 | 0.09 |
ENSMUST00000232294.2
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr1_+_74430575 | 0.08 |
ENSMUST00000027367.14
|
Ctdsp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr15_-_83479312 | 0.08 |
ENSMUST00000016901.5
|
Ttll12
|
tubulin tyrosine ligase-like family, member 12 |
| chr5_-_77053226 | 0.08 |
ENSMUST00000135954.2
|
Aasdh
|
aminoadipate-semialdehyde dehydrogenase |
| chr5_+_90666791 | 0.08 |
ENSMUST00000113179.9
ENSMUST00000128740.2 |
Afm
|
afamin |
| chr17_-_45996899 | 0.08 |
ENSMUST00000145873.8
|
Tmem63b
|
transmembrane protein 63b |
| chr5_-_121665249 | 0.08 |
ENSMUST00000152270.8
|
Mapkapk5
|
MAP kinase-activated protein kinase 5 |
| chr7_+_51537645 | 0.08 |
ENSMUST00000208711.2
|
Gas2
|
growth arrest specific 2 |
| chr3_-_148696155 | 0.08 |
ENSMUST00000196526.5
ENSMUST00000200543.5 ENSMUST00000200154.5 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr5_-_144160397 | 0.08 |
ENSMUST00000085701.7
|
Tecpr1
|
tectonin beta-propeller repeat containing 1 |
| chr1_-_85239791 | 0.08 |
ENSMUST00000162421.2
|
C130026I21Rik
|
RIKEN cDNA C130026I21 gene |
| chr8_+_34007333 | 0.08 |
ENSMUST00000124496.8
|
Tex15
|
testis expressed gene 15 |
| chr2_+_85804239 | 0.07 |
ENSMUST00000217244.2
|
Olfr1029
|
olfactory receptor 1029 |
| chr7_+_26932425 | 0.07 |
ENSMUST00000003860.13
ENSMUST00000108378.10 |
Coq8b
|
coenzyme Q8B |
| chr4_+_149188585 | 0.07 |
ENSMUST00000103216.10
ENSMUST00000030816.4 |
Dffa
|
DNA fragmentation factor, alpha subunit |
| chr11_-_5657658 | 0.07 |
ENSMUST00000154330.2
|
Mrps24
|
mitochondrial ribosomal protein S24 |
| chr6_-_118396321 | 0.07 |
ENSMUST00000032237.8
|
Bms1
|
BMS1, ribosome biogenesis factor |
| chr1_-_120198804 | 0.07 |
ENSMUST00000112641.8
|
Steap3
|
STEAP family member 3 |
| chr9_-_71803354 | 0.07 |
ENSMUST00000184448.8
|
Tcf12
|
transcription factor 12 |
| chr12_+_84463970 | 0.07 |
ENSMUST00000183146.2
|
Rnf113a2
|
ring finger protein 113A2 |
| chr9_-_113537277 | 0.06 |
ENSMUST00000111861.4
ENSMUST00000035086.13 |
Pdcd6ip
|
programmed cell death 6 interacting protein |
| chr9_-_82856321 | 0.06 |
ENSMUST00000189985.2
|
Phip
|
pleckstrin homology domain interacting protein |
| chr16_-_87229367 | 0.06 |
ENSMUST00000232095.2
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chr7_-_109585649 | 0.06 |
ENSMUST00000094097.12
|
Tmem41b
|
transmembrane protein 41B |
| chr2_-_79259235 | 0.06 |
ENSMUST00000143974.2
|
Cerkl
|
ceramide kinase-like |
| chr10_-_127358300 | 0.06 |
ENSMUST00000026470.6
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr5_-_134258435 | 0.06 |
ENSMUST00000016094.13
ENSMUST00000111275.8 ENSMUST00000144086.2 |
Ncf1
|
neutrophil cytosolic factor 1 |
| chr19_+_4008645 | 0.06 |
ENSMUST00000179433.8
|
Aldh3b3
|
aldehyde dehydrogenase 3 family, member B3 |
| chr10_-_81200680 | 0.06 |
ENSMUST00000131736.8
|
4930404N11Rik
|
RIKEN cDNA 4930404N11 gene |
| chr1_+_171246593 | 0.06 |
ENSMUST00000171362.2
|
Tstd1
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
| chr12_+_65272495 | 0.06 |
ENSMUST00000221980.2
|
Wdr20rt
|
WD repeat domain 20, retrogene |
| chr4_-_94444975 | 0.06 |
ENSMUST00000030313.9
|
Caap1
|
caspase activity and apoptosis inhibitor 1 |
| chr11_-_100504159 | 0.06 |
ENSMUST00000146840.3
|
Dnajc7
|
DnaJ heat shock protein family (Hsp40) member C7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb16
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0032379 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.2 | 0.7 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.1 | 0.7 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 0.4 | GO:2000566 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.1 | 0.6 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.1 | 0.4 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.1 | 0.6 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.3 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 0.4 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.1 | 0.3 | GO:0030887 | positive regulation of myeloid dendritic cell activation(GO:0030887) |
| 0.1 | 0.4 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 0.4 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 0.8 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.2 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.3 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.1 | 0.8 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.3 | GO:0072737 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.1 | 0.3 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.1 | 0.3 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.3 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.7 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 0.1 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.2 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.1 | 0.7 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.2 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.0 | 0.5 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.0 | 0.2 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) oxygen metabolic process(GO:0072592) |
| 0.0 | 0.4 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.4 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.3 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.4 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.2 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.2 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.8 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.5 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.2 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.1 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.0 | 0.1 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.3 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.3 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.1 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.0 | 0.6 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.6 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.2 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.3 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.0 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.0 | 0.1 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.0 | 0.1 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.0 | 0.3 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.2 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.0 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) bile acid signaling pathway(GO:0038183) |
| 0.0 | 0.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.6 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 0.2 | GO:0044753 | amphisome(GO:0044753) |
| 0.1 | 0.4 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.2 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 0.3 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 0.9 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 0.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.6 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.4 | GO:0001652 | granular component(GO:0001652) box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.3 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.7 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.3 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.6 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.5 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.7 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.5 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0071010 | prespliceosome(GO:0071010) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0050632 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.2 | 0.6 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.1 | 0.4 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.6 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.1 | 0.3 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.1 | 0.3 | GO:0032394 | MHC class Ib receptor activity(GO:0032394) |
| 0.1 | 0.4 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 0.3 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 0.6 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.9 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.2 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.2 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.1 | 0.2 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 0.8 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.1 | 0.2 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.1 | 0.7 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.5 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.3 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 0.3 | GO:0004043 | L-aminoadipate-semialdehyde dehydrogenase activity(GO:0004043) |
| 0.0 | 0.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.4 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.0 | 0.4 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.3 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.3 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.4 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.2 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.3 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.4 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.0 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.0 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.0 | 0.0 | GO:1902271 | lithocholic acid binding(GO:1902121) D3 vitamins binding(GO:1902271) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.2 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.3 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.7 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.0 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.8 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.6 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 1.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |