Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
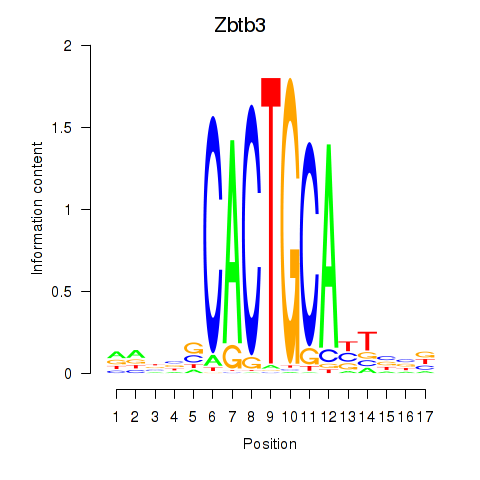
Results for Zbtb3
Z-value: 0.78
Transcription factors associated with Zbtb3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb3
|
ENSMUSG00000071661.8 | zinc finger and BTB domain containing 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb3 | mm39_v1_chr19_+_8779903_8779922 | -0.92 | 2.7e-02 | Click! |
Activity profile of Zbtb3 motif
Sorted Z-values of Zbtb3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_3551003 | 0.68 |
ENSMUST00000065703.9
ENSMUST00000203020.3 ENSMUST00000203821.3 |
Tarm1
|
T cell-interacting, activating receptor on myeloid cells 1 |
| chr2_-_89855921 | 0.68 |
ENSMUST00000216616.3
|
Olfr1264
|
olfactory receptor 1264 |
| chr7_-_114162125 | 0.66 |
ENSMUST00000211506.2
ENSMUST00000119712.8 ENSMUST00000032908.15 |
Cyp2r1
|
cytochrome P450, family 2, subfamily r, polypeptide 1 |
| chr1_+_132973724 | 0.44 |
ENSMUST00000077730.7
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
| chr11_-_120622770 | 0.39 |
ENSMUST00000154565.2
ENSMUST00000026148.9 |
Cbr2
|
carbonyl reductase 2 |
| chr10_+_60182630 | 0.39 |
ENSMUST00000020301.14
ENSMUST00000105460.8 ENSMUST00000170507.8 |
Vsir
|
V-set immunoregulatory receptor |
| chrX_-_166047289 | 0.35 |
ENSMUST00000133722.2
|
Tlr8
|
toll-like receptor 8 |
| chr9_+_21746785 | 0.31 |
ENSMUST00000058777.8
|
Angptl8
|
angiopoietin-like 8 |
| chr6_+_57183497 | 0.30 |
ENSMUST00000227298.2
|
Vmn1r13
|
vomeronasal 1 receptor 13 |
| chr9_-_89505178 | 0.29 |
ENSMUST00000044491.13
|
Minar1
|
membrane integral NOTCH2 associated receptor 1 |
| chr14_+_62793175 | 0.28 |
ENSMUST00000039064.8
|
Fam124a
|
family with sequence similarity 124, member A |
| chr19_-_4384029 | 0.26 |
ENSMUST00000176653.2
|
Kdm2a
|
lysine (K)-specific demethylase 2A |
| chr15_-_66985760 | 0.25 |
ENSMUST00000092640.6
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr4_+_40269563 | 0.25 |
ENSMUST00000129758.3
|
Smim27
|
small integral membrane protein 27 |
| chr9_+_107458495 | 0.24 |
ENSMUST00000040059.9
|
Hyal3
|
hyaluronoglucosaminidase 3 |
| chr8_-_123980908 | 0.23 |
ENSMUST00000122363.8
|
Vps9d1
|
VPS9 domain containing 1 |
| chr2_+_113158100 | 0.23 |
ENSMUST00000102547.10
|
Fmn1
|
formin 1 |
| chr18_-_16942289 | 0.22 |
ENSMUST00000025166.14
|
Cdh2
|
cadherin 2 |
| chr2_-_25517945 | 0.22 |
ENSMUST00000028307.9
|
Fcna
|
ficolin A |
| chrX_-_166047275 | 0.22 |
ENSMUST00000112170.2
|
Tlr8
|
toll-like receptor 8 |
| chr8_+_65399831 | 0.20 |
ENSMUST00000026595.13
ENSMUST00000209852.2 ENSMUST00000079896.9 |
Tmem192
|
transmembrane protein 192 |
| chr14_+_66378382 | 0.20 |
ENSMUST00000022620.11
|
Chrna2
|
cholinergic receptor, nicotinic, alpha polypeptide 2 (neuronal) |
| chr14_-_19261196 | 0.19 |
ENSMUST00000112797.12
ENSMUST00000225885.2 |
D830030K20Rik
|
RIKEN cDNA D830030K20 gene |
| chr9_-_40442669 | 0.18 |
ENSMUST00000119373.9
|
Gramd1b
|
GRAM domain containing 1B |
| chr11_+_92989229 | 0.17 |
ENSMUST00000107859.8
ENSMUST00000107861.8 ENSMUST00000042943.13 ENSMUST00000107858.9 |
Car10
|
carbonic anhydrase 10 |
| chr11_-_102076028 | 0.16 |
ENSMUST00000107156.9
ENSMUST00000021297.6 |
Lsm12
|
LSM12 homolog |
| chr10_+_74802996 | 0.16 |
ENSMUST00000037813.5
|
Gnaz
|
guanine nucleotide binding protein, alpha z subunit |
| chr17_-_37523969 | 0.15 |
ENSMUST00000060728.7
ENSMUST00000216318.2 |
Olfr95
|
olfactory receptor 95 |
| chr8_-_3517617 | 0.15 |
ENSMUST00000111081.10
ENSMUST00000004686.13 |
Pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr19_+_10366450 | 0.15 |
ENSMUST00000073899.6
|
Syt7
|
synaptotagmin VII |
| chr9_-_118917275 | 0.14 |
ENSMUST00000214470.2
|
Plcd1
|
phospholipase C, delta 1 |
| chr2_+_172235702 | 0.14 |
ENSMUST00000099061.9
ENSMUST00000103073.9 |
Cass4
|
Cas scaffolding protein family member 4 |
| chr14_-_20027279 | 0.14 |
ENSMUST00000160013.8
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr11_+_115921129 | 0.14 |
ENSMUST00000021116.12
ENSMUST00000106452.2 |
Unk
|
unkempt family zinc finger |
| chr10_+_79766254 | 0.13 |
ENSMUST00000131118.2
|
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr7_-_44498305 | 0.13 |
ENSMUST00000207293.2
ENSMUST00000207532.2 |
Tbc1d17
|
TBC1 domain family, member 17 |
| chr4_-_132260799 | 0.13 |
ENSMUST00000152993.8
ENSMUST00000067496.7 |
Atpif1
|
ATPase inhibitory factor 1 |
| chr17_+_35235552 | 0.13 |
ENSMUST00000007245.8
ENSMUST00000172499.2 |
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr14_+_54032814 | 0.13 |
ENSMUST00000103671.4
|
Trav13-5
|
T cell receptor alpha variable 13-5 |
| chr7_-_80037688 | 0.12 |
ENSMUST00000080932.8
|
Fes
|
feline sarcoma oncogene |
| chr11_-_72380730 | 0.12 |
ENSMUST00000045303.10
|
Spns2
|
spinster homolog 2 |
| chr15_+_102052797 | 0.12 |
ENSMUST00000023807.7
|
Igfbp6
|
insulin-like growth factor binding protein 6 |
| chr5_+_146885450 | 0.12 |
ENSMUST00000146511.8
ENSMUST00000132102.2 |
Gtf3a
|
general transcription factor III A |
| chr17_+_24768808 | 0.12 |
ENSMUST00000228550.2
ENSMUST00000035565.5 |
Pkd1
|
polycystin 1, transient receptor poteintial channel interacting |
| chr6_+_72281587 | 0.11 |
ENSMUST00000183018.8
ENSMUST00000182014.10 |
Sftpb
|
surfactant associated protein B |
| chr5_+_138083345 | 0.11 |
ENSMUST00000019660.11
ENSMUST00000066617.12 ENSMUST00000110963.8 |
Zkscan1
|
zinc finger with KRAB and SCAN domains 1 |
| chr2_-_119378108 | 0.11 |
ENSMUST00000060009.9
|
Exd1
|
exonuclease 3'-5' domain containing 1 |
| chr15_-_82223065 | 0.11 |
ENSMUST00000229733.2
ENSMUST00000229388.2 |
Naga
|
N-acetyl galactosaminidase, alpha |
| chr11_+_98700613 | 0.11 |
ENSMUST00000169695.2
|
Casc3
|
cancer susceptibility candidate 3 |
| chr9_+_21527526 | 0.11 |
ENSMUST00000174008.8
|
Smarca4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr9_+_44410417 | 0.11 |
ENSMUST00000074989.7
ENSMUST00000218913.2 |
Bcl9l
|
B cell CLL/lymphoma 9-like |
| chr7_+_141276575 | 0.10 |
ENSMUST00000185406.8
|
Muc2
|
mucin 2 |
| chr6_+_134897364 | 0.10 |
ENSMUST00000067327.11
ENSMUST00000003115.9 |
Cdkn1b
|
cyclin-dependent kinase inhibitor 1B |
| chr3_-_115800989 | 0.10 |
ENSMUST00000067485.4
|
Slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr8_-_70892204 | 0.10 |
ENSMUST00000076615.6
|
Crtc1
|
CREB regulated transcription coactivator 1 |
| chr9_+_21527462 | 0.09 |
ENSMUST00000034707.15
ENSMUST00000098948.10 |
Smarca4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr5_-_5744559 | 0.09 |
ENSMUST00000115426.9
|
Steap2
|
six transmembrane epithelial antigen of prostate 2 |
| chr7_+_28466658 | 0.09 |
ENSMUST00000155327.8
|
Sirt2
|
sirtuin 2 |
| chrX_-_72380446 | 0.09 |
ENSMUST00000207943.2
|
Gm45015
|
predicted gene 45015 |
| chr1_-_162687488 | 0.08 |
ENSMUST00000134098.8
ENSMUST00000111518.3 |
Fmo1
|
flavin containing monooxygenase 1 |
| chr14_+_36789999 | 0.08 |
ENSMUST00000057176.5
|
Lrit2
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 |
| chr9_+_92157799 | 0.08 |
ENSMUST00000126911.2
|
Plscr2
|
phospholipid scramblase 2 |
| chr13_-_96678844 | 0.08 |
ENSMUST00000223475.2
|
Polk
|
polymerase (DNA directed), kappa |
| chr7_+_18758289 | 0.08 |
ENSMUST00000023882.14
ENSMUST00000153976.8 |
Sympk
|
symplekin |
| chr7_-_80037622 | 0.08 |
ENSMUST00000206698.2
|
Fes
|
feline sarcoma oncogene |
| chr3_-_37286714 | 0.07 |
ENSMUST00000161015.2
ENSMUST00000029273.8 |
Il21
|
interleukin 21 |
| chr5_-_45796857 | 0.07 |
ENSMUST00000016023.9
|
Fam184b
|
family with sequence similarity 184, member B |
| chr18_+_34380738 | 0.07 |
ENSMUST00000066133.7
|
Apc
|
APC, WNT signaling pathway regulator |
| chr2_+_113158053 | 0.07 |
ENSMUST00000099576.9
|
Fmn1
|
formin 1 |
| chr9_-_45896110 | 0.06 |
ENSMUST00000215060.2
ENSMUST00000213853.2 ENSMUST00000216334.2 |
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr2_+_74566740 | 0.06 |
ENSMUST00000111982.8
|
Hoxd3
|
homeobox D3 |
| chr15_+_103181311 | 0.06 |
ENSMUST00000100162.5
ENSMUST00000230893.2 |
Copz1
|
coatomer protein complex, subunit zeta 1 |
| chr3_+_79793237 | 0.06 |
ENSMUST00000029567.9
|
Gask1b
|
golgi associated kinase 1B |
| chr7_-_110681402 | 0.06 |
ENSMUST00000159305.2
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr19_-_8382424 | 0.05 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr13_+_76727799 | 0.05 |
ENSMUST00000109589.3
|
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr11_-_16952929 | 0.05 |
ENSMUST00000156101.2
|
Plek
|
pleckstrin |
| chr14_+_53994813 | 0.05 |
ENSMUST00000180380.3
|
Trav13-4-dv7
|
T cell receptor alpha variable 13-4-DV7 |
| chr5_-_90371816 | 0.04 |
ENSMUST00000118816.6
ENSMUST00000048363.10 |
Cox18
|
cytochrome c oxidase assembly protein 18 |
| chr6_-_28421678 | 0.04 |
ENSMUST00000090511.4
|
Gcc1
|
golgi coiled coil 1 |
| chr11_+_87482971 | 0.04 |
ENSMUST00000103179.10
ENSMUST00000092802.12 ENSMUST00000146871.8 |
Mtmr4
|
myotubularin related protein 4 |
| chr5_+_76988444 | 0.04 |
ENSMUST00000120639.9
ENSMUST00000163347.8 ENSMUST00000121851.2 |
Cracd
|
capping protein inhibiting regulator of actin |
| chrX_-_161747552 | 0.03 |
ENSMUST00000038769.3
|
S100g
|
S100 calcium binding protein G |
| chr7_-_118594365 | 0.03 |
ENSMUST00000008878.10
|
Gprc5b
|
G protein-coupled receptor, family C, group 5, member B |
| chr5_-_39034039 | 0.03 |
ENSMUST00000169819.5
ENSMUST00000171633.5 |
Clnk
|
cytokine-dependent hematopoietic cell linker |
| chr3_+_107198528 | 0.03 |
ENSMUST00000029502.14
|
Slc16a4
|
solute carrier family 16 (monocarboxylic acid transporters), member 4 |
| chr18_+_90528308 | 0.03 |
ENSMUST00000235634.2
|
Tmx3
|
thioredoxin-related transmembrane protein 3 |
| chr10_+_128104525 | 0.03 |
ENSMUST00000050901.5
|
Apof
|
apolipoprotein F |
| chr9_+_44990447 | 0.03 |
ENSMUST00000050020.8
|
Jaml
|
junction adhesion molecule like |
| chr1_-_121255753 | 0.03 |
ENSMUST00000003818.14
|
Insig2
|
insulin induced gene 2 |
| chr16_+_34511073 | 0.03 |
ENSMUST00000231609.2
|
Ccdc14
|
coiled-coil domain containing 14 |
| chr5_+_72805122 | 0.03 |
ENSMUST00000087212.8
|
Nipal1
|
NIPA-like domain containing 1 |
| chr19_+_10366753 | 0.02 |
ENSMUST00000169121.9
ENSMUST00000076968.11 ENSMUST00000235479.2 ENSMUST00000223586.2 ENSMUST00000235784.2 ENSMUST00000224135.3 ENSMUST00000225452.3 ENSMUST00000237366.2 |
Syt7
|
synaptotagmin VII |
| chr9_+_44990502 | 0.02 |
ENSMUST00000216426.2
|
Jaml
|
junction adhesion molecule like |
| chr16_+_11223512 | 0.02 |
ENSMUST00000096273.9
|
Snx29
|
sorting nexin 29 |
| chr1_+_156138286 | 0.02 |
ENSMUST00000027896.10
|
Nphs2
|
nephrosis 2, podocin |
| chr1_-_133849131 | 0.02 |
ENSMUST00000048432.6
|
Prelp
|
proline arginine-rich end leucine-rich repeat |
| chr6_-_128868068 | 0.02 |
ENSMUST00000178918.2
ENSMUST00000160290.8 |
BC035044
|
cDNA sequence BC035044 |
| chr2_+_80447389 | 0.02 |
ENSMUST00000028384.5
|
Dusp19
|
dual specificity phosphatase 19 |
| chr9_-_45896075 | 0.02 |
ENSMUST00000217636.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr1_-_121255503 | 0.02 |
ENSMUST00000160688.2
|
Insig2
|
insulin induced gene 2 |
| chr18_-_80512850 | 0.02 |
ENSMUST00000036229.13
|
Ctdp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr13_+_96679110 | 0.01 |
ENSMUST00000179226.8
|
Cert1
|
ceramide transporter 1 |
| chr2_-_35226981 | 0.01 |
ENSMUST00000028241.7
|
Stom
|
stomatin |
| chr1_-_121255448 | 0.01 |
ENSMUST00000186915.2
ENSMUST00000160968.8 ENSMUST00000162582.2 |
Insig2
|
insulin induced gene 2 |
| chr12_-_113552322 | 0.01 |
ENSMUST00000103443.3
|
Ighv2-2
|
immunoglobulin heavy variable 2-2 |
| chrX_+_165021897 | 0.01 |
ENSMUST00000112235.8
|
Gpm6b
|
glycoprotein m6b |
| chr1_-_172085977 | 0.01 |
ENSMUST00000111243.2
|
Atp1a4
|
ATPase, Na+/K+ transporting, alpha 4 polypeptide |
| chr19_+_25214322 | 0.01 |
ENSMUST00000049400.15
|
Kank1
|
KN motif and ankyrin repeat domains 1 |
| chrX_-_36990581 | 0.01 |
ENSMUST00000170643.4
ENSMUST00000089062.8 |
Rhox9
|
reproductive homeobox 9 |
| chr12_+_112455882 | 0.01 |
ENSMUST00000057465.7
ENSMUST00000223266.2 |
A530016L24Rik
|
RIKEN cDNA A530016L24 gene |
| chr15_-_77639466 | 0.01 |
ENSMUST00000089450.5
|
Apol8
|
apolipoprotein L 8 |
| chr13_-_95359543 | 0.01 |
ENSMUST00000162292.8
|
Pde8b
|
phosphodiesterase 8B |
| chr8_+_84682136 | 0.01 |
ENSMUST00000005607.9
|
Asf1b
|
anti-silencing function 1B histone chaperone |
| chr3_+_115801106 | 0.01 |
ENSMUST00000029575.12
ENSMUST00000106501.8 |
Extl2
|
exostosin-like glycosyltransferase 2 |
| chr7_+_101555111 | 0.01 |
ENSMUST00000033131.12
ENSMUST00000193465.2 |
Lamtor1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr14_+_53995108 | 0.01 |
ENSMUST00000184905.2
|
Trav13-4-dv7
|
T cell receptor alpha variable 13-4-DV7 |
| chr14_+_53088747 | 0.01 |
ENSMUST00000103588.4
|
Trav13d-1
|
T cell receptor alpha variable 13D-1 |
| chr13_-_96678987 | 0.01 |
ENSMUST00000022172.12
|
Polk
|
polymerase (DNA directed), kappa |
| chr16_+_34510918 | 0.01 |
ENSMUST00000023532.7
|
Ccdc14
|
coiled-coil domain containing 14 |
| chrX_+_52609954 | 0.01 |
ENSMUST00000063384.4
|
Rtl8c
|
retrotransposon Gag like 8C |
| chr14_+_53180221 | 0.01 |
ENSMUST00000197954.2
|
Trav13d-2
|
T cell receptor alpha variable 13D-2 |
| chr2_+_152404897 | 0.01 |
ENSMUST00000238626.2
|
Gm17416
|
predicted gene, 17416 |
| chr9_+_46180362 | 0.01 |
ENSMUST00000214202.2
ENSMUST00000215458.2 ENSMUST00000215187.2 ENSMUST00000213878.2 ENSMUST00000034584.4 |
Apoa5
|
apolipoprotein A-V |
| chr14_+_53574579 | 0.01 |
ENSMUST00000179580.3
|
Trav13n-3
|
T cell receptor alpha variable 13N-3 |
| chr10_-_108535970 | 0.01 |
ENSMUST00000218023.2
|
Gm5136
|
predicted gene 5136 |
| chr4_-_43700807 | 0.01 |
ENSMUST00000055545.5
|
Olfr70
|
olfactory receptor 70 |
| chr15_+_92294762 | 0.01 |
ENSMUST00000169942.10
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr14_+_53270305 | 0.01 |
ENSMUST00000179512.3
|
Trav13d-3
|
T cell receptor alpha variable 13D-3 |
| chr10_+_76089674 | 0.01 |
ENSMUST00000036387.8
|
S100b
|
S100 protein, beta polypeptide, neural |
| chr11_-_48792973 | 0.01 |
ENSMUST00000109210.8
|
Gm5431
|
predicted gene 5431 |
| chr14_+_53601293 | 0.01 |
ENSMUST00000103634.3
|
Trav13n-4
|
T cell receptor alpha variable 13N-4 |
| chr9_+_92157655 | 0.01 |
ENSMUST00000034932.14
ENSMUST00000180154.8 |
Plscr2
|
phospholipid scramblase 2 |
| chr10_+_79500421 | 0.01 |
ENSMUST00000217748.2
|
Madcam1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chr5_+_72805153 | 0.01 |
ENSMUST00000197837.3
|
Nipal1
|
NIPA-like domain containing 1 |
| chr10_-_77618376 | 0.00 |
ENSMUST00000167358.2
|
Gm3250
|
predicted gene 3250 |
| chr1_-_162687254 | 0.00 |
ENSMUST00000131058.8
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr2_-_180798785 | 0.00 |
ENSMUST00000055990.8
|
Eef1a2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr16_+_20408886 | 0.00 |
ENSMUST00000232279.2
ENSMUST00000232474.2 |
Vwa5b2
|
von Willebrand factor A domain containing 5B2 |
| chrX_+_160080789 | 0.00 |
ENSMUST00000207853.2
ENSMUST00000207800.2 |
Gm15262
|
predicted gene 15262 |
| chr10_+_79500387 | 0.00 |
ENSMUST00000020554.8
|
Madcam1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chr7_-_62862261 | 0.00 |
ENSMUST00000032738.7
|
Chrna7
|
cholinergic receptor, nicotinic, alpha polypeptide 7 |
| chr1_-_162687369 | 0.00 |
ENSMUST00000193078.6
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr5_-_142495408 | 0.00 |
ENSMUST00000110784.8
|
Radil
|
Ras association and DIL domains |
| chr6_-_136834725 | 0.00 |
ENSMUST00000032341.3
|
Art4
|
ADP-ribosyltransferase 4 |
| chr7_-_134100659 | 0.00 |
ENSMUST00000238294.2
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed |
| chr14_+_53180476 | 0.00 |
ENSMUST00000103596.3
|
Trav13d-2
|
T cell receptor alpha variable 13D-2 |
| chr5_+_43819688 | 0.00 |
ENSMUST00000048150.15
|
Cc2d2a
|
coiled-coil and C2 domain containing 2A |
| chr10_-_62723103 | 0.00 |
ENSMUST00000218438.2
|
Tet1
|
tet methylcytosine dioxygenase 1 |
| chr11_-_103511966 | 0.00 |
ENSMUST00000059279.13
|
Gm884
|
predicted gene 884 |
| chr15_-_77639418 | 0.00 |
ENSMUST00000229445.2
|
Apol8
|
apolipoprotein L 8 |
| chr18_+_44467133 | 0.00 |
ENSMUST00000025349.12
ENSMUST00000115498.2 |
Myot
|
myotilin |
| chr16_-_10609959 | 0.00 |
ENSMUST00000037996.7
|
Prm2
|
protamine 2 |
| chr14_+_53491249 | 0.00 |
ENSMUST00000196941.2
|
Trav13n-2
|
T cell receptor alpha variable 13N-2 |
| chrX_+_160080763 | 0.00 |
ENSMUST00000207353.2
|
Gm15262
|
predicted gene 15262 |
| chr7_-_119744509 | 0.00 |
ENSMUST00000208874.2
ENSMUST00000033207.6 |
Zp2
|
zona pellucida glycoprotein 2 |
| chr6_+_18170686 | 0.00 |
ENSMUST00000045706.12
|
Cftr
|
cystic fibrosis transmembrane conductance regulator |
| chr7_-_103661957 | 0.00 |
ENSMUST00000106862.3
|
Olfr639
|
olfactory receptor 639 |
| chr11_+_109304735 | 0.00 |
ENSMUST00000055404.8
|
9930022D16Rik
|
RIKEN cDNA 9930022D16 gene |
| chr18_-_3299452 | 0.00 |
ENSMUST00000126578.8
|
Crem
|
cAMP responsive element modulator |
| chr14_-_20027219 | 0.00 |
ENSMUST00000055100.14
ENSMUST00000162425.8 |
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chrX_+_106192510 | 0.00 |
ENSMUST00000147521.8
ENSMUST00000167673.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr9_-_121620150 | 0.00 |
ENSMUST00000215910.2
ENSMUST00000215477.2 ENSMUST00000163981.3 |
Hhatl
|
hedgehog acyltransferase-like |
| chr19_-_8109346 | 0.00 |
ENSMUST00000065651.5
|
Slc22a28
|
solute carrier family 22, member 28 |
| chr5_+_89675288 | 0.00 |
ENSMUST00000048557.3
|
Npffr2
|
neuropeptide FF receptor 2 |
| chr10_-_62723238 | 0.00 |
ENSMUST00000228901.2
|
Tet1
|
tet methylcytosine dioxygenase 1 |
| chr14_+_53399856 | 0.00 |
ENSMUST00000198359.2
|
Trav13n-1
|
T cell receptor alpha variable 13N-1 |
| chr9_-_40873629 | 0.00 |
ENSMUST00000160120.2
|
Jhy
|
junctional cadherin complex regulator |
| chr13_+_68011442 | 0.00 |
ENSMUST00000078471.7
|
BC048507
|
cDNA sequence BC048507 |
| chr16_+_22739191 | 0.00 |
ENSMUST00000116625.10
|
Fetub
|
fetuin beta |
| chr2_-_127634387 | 0.00 |
ENSMUST00000135091.2
|
Mtln
|
mitoregulin |
| chr13_-_95359955 | 0.00 |
ENSMUST00000159608.8
|
Pde8b
|
phosphodiesterase 8B |
| chr5_+_33261563 | 0.00 |
ENSMUST00000011178.5
|
Slc5a1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.6 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 0.4 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.1 | 0.2 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.1 | 0.4 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 0.3 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.3 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.0 | 0.2 | GO:1902661 | DNA methylation on cytosine within a CG sequence(GO:0010424) regulation of glucose mediated signaling pathway(GO:1902659) positive regulation of glucose mediated signaling pathway(GO:1902661) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.7 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.2 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.3 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 0.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.1 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.0 | GO:0010924 | regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010924) positive regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010925) phospholipase C-inhibiting G-protein coupled receptor signaling pathway(GO:0030845) regulation of cell diameter(GO:0060305) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) eye pigmentation(GO:0048069) |
| 0.0 | 0.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.4 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0008456 | alpha-N-acetylgalactosaminidase activity(GO:0008456) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.2 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |