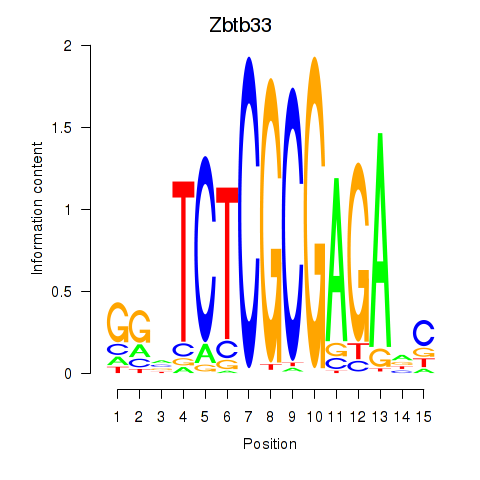
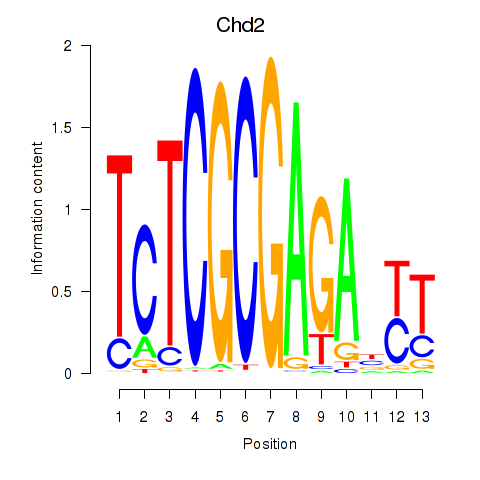
chr12_+_30634425
Show fit
1.78
ENSMUST00000057151.10
Tmem18
transmembrane protein 18
chr5_+_124045238
Show fit
1.47
ENSMUST00000023869.15
Denr
density-regulated protein
chr17_+_28910302
Show fit
1.44
ENSMUST00000004990.14
ENSMUST00000114754.8
ENSMUST00000062694.16
Mapk14
mitogen-activated protein kinase 14
chr5_+_135807334
Show fit
1.43
ENSMUST00000019323.11
Mdh2
malate dehydrogenase 2, NAD (mitochondrial)
chr9_-_57375269
Show fit
1.42
ENSMUST00000215059.2
ENSMUST00000046587.8
ENSMUST00000214256.2
Scamp5
secretory carrier membrane protein 5
chr5_+_140307369
Show fit
1.35
ENSMUST00000198660.5
Nudt1
nudix (nucleoside diphosphate linked moiety X)-type motif 1
chr7_-_117715351
Show fit
1.32
ENSMUST00000128482.8
ENSMUST00000131840.3
Rps15a
ribosomal protein S15A
chr11_-_70537690
Show fit
1.31
ENSMUST00000136383.2
Slc25a11
solute carrier family 25 (mitochondrial carrier oxoglutarate carrier), member 11
chr5_+_135807528
Show fit
1.24
ENSMUST00000200556.5
ENSMUST00000196285.2
Mdh2
malate dehydrogenase 2, NAD (mitochondrial)
chr17_-_46558894
Show fit
1.23
ENSMUST00000142706.9
ENSMUST00000173349.8
ENSMUST00000087026.13
Polr1c
polymerase (RNA) I polypeptide C
chr4_+_116543045
Show fit
1.22
ENSMUST00000129315.8
ENSMUST00000106470.8
Prdx1
peroxiredoxin 1
chr3_+_105866685
Show fit
1.20
ENSMUST00000010278.12
Wdr77
WD repeat domain 77
chr7_-_112968533
Show fit
1.19
ENSMUST00000047091.14
ENSMUST00000119278.8
Btbd10
BTB (POZ) domain containing 10
chr4_+_86493905
Show fit
1.10
ENSMUST00000091064.8
Rraga
Ras-related GTP binding A
chr6_-_148113494
Show fit
1.10
ENSMUST00000126698.2
Ergic2
ERGIC and golgi 2
chr5_+_124045552
Show fit
1.06
ENSMUST00000166233.2
Denr
density-regulated protein
chr1_+_160022785
Show fit
1.00
ENSMUST00000135680.8
ENSMUST00000097193.3
Mrps14
mitochondrial ribosomal protein S14
chr15_-_98660873
Show fit
0.92
ENSMUST00000156572.3
Arf3
ADP-ribosylation factor 3
chr9_-_108455899
Show fit
0.92
ENSMUST00000068700.7
Wdr6
WD repeat domain 6
chr3_+_88243562
Show fit
0.91
ENSMUST00000001451.11
Smg5
Smg-5 homolog, nonsense mediated mRNA decay factor (C. elegans)
chr10_-_119075910
Show fit
0.89
ENSMUST00000020315.13
Cand1
Links
cullin associated and neddylation disassociated 1
chr12_+_82216193
Show fit
0.88
ENSMUST00000166429.9
ENSMUST00000220963.2
Sipa1l1
signal-induced proliferation-associated 1 like 1
chr17_+_28910393
Show fit
0.88
ENSMUST00000124886.9
ENSMUST00000114758.9
Mapk14
mitogen-activated protein kinase 14
chr9_-_55419442
Show fit
0.86
ENSMUST00000034866.9
Etfa
electron transferring flavoprotein, alpha polypeptide
chr11_+_74661647
Show fit
0.86
ENSMUST00000010698.13
ENSMUST00000141755.8
Mettl16
methyltransferase like 16
chr9_-_64080161
Show fit
0.85
ENSMUST00000176299.8
ENSMUST00000130127.8
ENSMUST00000176794.8
ENSMUST00000177045.8
Zwilch
zwilch kinetochore protein
chr8_-_123939480
Show fit
0.84
ENSMUST00000000759.9
Chmp1a
charged multivesicular body protein 1A
chr2_-_34760960
Show fit
0.82
ENSMUST00000028225.12
Psmd5
proteasome (prosome, macropain) 26S subunit, non-ATPase, 5
chr1_+_58432629
Show fit
0.79
ENSMUST00000186949.2
Bzw1
basic leucine zipper and W2 domains 1
chr11_-_34724458
Show fit
0.79
ENSMUST00000093191.3
Spdl1
spindle apparatus coiled-coil protein 1
chr11_-_119190830
Show fit
0.78
ENSMUST00000106253.2
Eif4a3
eukaryotic translation initiation factor 4A3
chr5_+_45827249
Show fit
0.77
ENSMUST00000117396.3
Ncapg
non-SMC condensin I complex, subunit G
chr8_-_46664321
Show fit
0.77
ENSMUST00000034049.5
Slc25a4
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4
chr8_+_53964721
Show fit
0.77
ENSMUST00000211424.2
ENSMUST00000033920.6
Aga
aspartylglucosaminidase
chr7_+_24584197
Show fit
0.77
ENSMUST00000156372.8
ENSMUST00000124035.2
Rps19
ribosomal protein S19
chr4_+_152123772
Show fit
0.76
ENSMUST00000084116.13
ENSMUST00000103197.5
Nol9
nucleolar protein 9
chr6_+_18848600
Show fit
0.75
ENSMUST00000201141.3
Lsm8
LSM8 homolog, U6 small nuclear RNA associated
chr11_+_90528953
Show fit
0.75
ENSMUST00000020851.15
Cox11
cytochrome c oxidase assembly protein 11, copper chaperone
chr2_-_26800581
Show fit
0.75
ENSMUST00000015920.12
ENSMUST00000139815.2
ENSMUST00000102899.10
Med22
mediator complex subunit 22
chr1_-_134289670
Show fit
0.72
ENSMUST00000049470.11
Tmem183a
transmembrane protein 183A
chr4_+_116542741
Show fit
0.72
ENSMUST00000135573.8
ENSMUST00000151129.8
Prdx1
peroxiredoxin 1
chr13_-_111626562
Show fit
0.72
ENSMUST00000091236.11
ENSMUST00000047627.14
Gpbp1
GC-rich promoter binding protein 1
chrX_+_105059305
Show fit
0.71
ENSMUST00000033582.5
Cox7b
cytochrome c oxidase subunit 7B
chr12_+_76812301
Show fit
0.70
ENSMUST00000041262.14
ENSMUST00000126408.2
ENSMUST00000110399.3
ENSMUST00000137826.8
Churc1
Fntb
churchill domain containing 1
chr8_-_26330468
Show fit
0.68
ENSMUST00000110609.8
Ash2l
ASH2 like histone lysine methyltransferase complex subunit
chr13_-_58549728
Show fit
0.68
ENSMUST00000225176.2
ENSMUST00000223822.2
Hnrnpk
heterogeneous nuclear ribonucleoprotein K
chr9_+_54771064
Show fit
0.67
ENSMUST00000034843.9
Ireb2
iron responsive element binding protein 2
chr11_-_70537878
Show fit
0.66
ENSMUST00000014750.15
Slc25a11
solute carrier family 25 (mitochondrial carrier oxoglutarate carrier), member 11
chr17_+_28910714
Show fit
0.66
ENSMUST00000233250.2
Mapk14
mitogen-activated protein kinase 14
chr8_-_95564881
Show fit
0.66
ENSMUST00000034233.15
ENSMUST00000162538.9
Ciapin1
cytokine induced apoptosis inhibitor 1
chr6_+_18848570
Show fit
0.65
ENSMUST00000056398.11
Lsm8
LSM8 homolog, U6 small nuclear RNA associated
chr8_-_120285457
Show fit
0.65
ENSMUST00000098362.11
ENSMUST00000081381.6
Mbtps1
membrane-bound transcription factor peptidase, site 1
chr8_-_23083751
Show fit
0.64
ENSMUST00000009036.11
Vdac3
voltage-dependent anion channel 3
chr11_-_119190896
Show fit
0.64
ENSMUST00000026667.15
Eif4a3
eukaryotic translation initiation factor 4A3
chr11_+_68961599
Show fit
0.63
ENSMUST00000075980.12
ENSMUST00000094081.5
Tmem107
transmembrane protein 107
chr4_+_135583018
Show fit
0.62
ENSMUST00000105853.10
ENSMUST00000097844.9
ENSMUST00000102544.9
ENSMUST00000126641.2
Srsf10
serine and arginine-rich splicing factor 10
chr6_-_148113872
Show fit
0.61
ENSMUST00000136008.8
Ergic2
ERGIC and golgi 2
chr5_-_117527094
Show fit
0.60
ENSMUST00000111953.2
ENSMUST00000086461.13
Rfc5
replication factor C (activator 1) 5
chr11_-_88755360
Show fit
0.59
ENSMUST00000018572.11
Akap1
A kinase (PRKA) anchor protein 1
chr13_+_81931196
Show fit
0.59
ENSMUST00000022009.10
ENSMUST00000223793.2
Cetn3
centrin 3
chr9_+_64080644
Show fit
0.58
ENSMUST00000034966.9
Rpl4
ribosomal protein L4
chr17_+_46558995
Show fit
0.58
ENSMUST00000095263.10
ENSMUST00000123311.8
Yipf3
Yip1 domain family, member 3
chr7_+_90091937
Show fit
0.55
ENSMUST00000061767.5
Crebzf
CREB/ATF bZIP transcription factor
chr17_+_24939037
Show fit
0.54
ENSMUST00000170715.8
Rps2
ribosomal protein S2
chr13_-_58550263
Show fit
0.54
ENSMUST00000177019.8
Hnrnpk
heterogeneous nuclear ribonucleoprotein K
chr4_+_24496434
Show fit
0.54
ENSMUST00000108222.9
ENSMUST00000138567.9
ENSMUST00000050446.13
Mms22l
MMS22-like, DNA repair protein
chr9_+_64188857
Show fit
0.53
ENSMUST00000215031.2
ENSMUST00000213165.2
ENSMUST00000213289.2
ENSMUST00000216594.2
ENSMUST00000034964.7
Tipin
timeless interacting protein
chr18_+_35695339
Show fit
0.53
ENSMUST00000237365.2
Matr3
matrin 3
chr3_+_151916059
Show fit
0.53
ENSMUST00000166984.8
Fubp1
far upstream element (FUSE) binding protein 1
chr10_-_85793639
Show fit
0.53
ENSMUST00000001834.4
Rtcb
RNA 2',3'-cyclic phosphate and 5'-OH ligase
chr10_-_127147609
Show fit
0.52
ENSMUST00000037290.12
ENSMUST00000171564.8
Mars1
methionine-tRNA synthetase 1
chr4_+_152381662
Show fit
0.51
ENSMUST00000048892.14
Icmt
isoprenylcysteine carboxyl methyltransferase
chr1_-_105591362
Show fit
0.50
ENSMUST00000187537.7
ENSMUST00000186485.7
ENSMUST00000190811.7
Pign
phosphatidylinositol glycan anchor biosynthesis, class N
chr2_-_144112700
Show fit
0.49
ENSMUST00000110030.10
Snx5
sorting nexin 5
chr8_-_23083829
Show fit
0.49
ENSMUST00000179233.2
Vdac3
voltage-dependent anion channel 3
chr16_-_87292592
Show fit
0.49
ENSMUST00000176750.2
ENSMUST00000175977.8
Cct8
chaperonin containing Tcp1, subunit 8 (theta)
chrX_+_13147209
Show fit
0.48
ENSMUST00000000804.7
Ddx3x
DEAD box helicase 3, X-linked
chr15_+_81756671
Show fit
0.48
ENSMUST00000135198.2
ENSMUST00000157003.8
ENSMUST00000229068.2
Aco2
aconitase 2, mitochondrial
chr13_+_81931642
Show fit
0.48
ENSMUST00000224574.2
Cetn3
centrin 3
chr4_-_118294521
Show fit
0.48
ENSMUST00000006565.13
Cdc20
cell division cycle 20
chr4_+_133302039
Show fit
0.48
ENSMUST00000030662.3
Gpatch3
G patch domain containing 3
chr4_-_134262509
Show fit
0.47
ENSMUST00000102550.10
Mtfr1l
mitochondrial fission regulator 1-like
chr7_-_34089109
Show fit
0.47
ENSMUST00000085585.12
Lsm14a
LSM14A mRNA processing body assembly factor
chr18_-_35064906
Show fit
0.46
ENSMUST00000025218.8
Etf1
eukaryotic translation termination factor 1
chr7_+_81412695
Show fit
0.46
ENSMUST00000133034.2
Ramac
RNA guanine-7 methyltransferase activating subunit
chr4_-_134262358
Show fit
0.46
ENSMUST00000154769.8
Mtfr1l
mitochondrial fission regulator 1-like
chr15_-_31601652
Show fit
0.46
ENSMUST00000161266.2
Cct5
chaperonin containing Tcp1, subunit 5 (epsilon)
chr11_+_115494751
Show fit
0.45
ENSMUST00000058109.9
Mrps7
mitchondrial ribosomal protein S7
chr5_-_110434026
Show fit
0.45
ENSMUST00000031472.12
Pxmp2
peroxisomal membrane protein 2
chr19_-_6117815
Show fit
0.44
ENSMUST00000162575.8
ENSMUST00000159084.8
ENSMUST00000161718.8
ENSMUST00000162810.8
ENSMUST00000025713.12
ENSMUST00000113543.9
ENSMUST00000160417.8
ENSMUST00000161528.2
Tm7sf2
transmembrane 7 superfamily member 2
chr2_-_131021905
Show fit
0.44
ENSMUST00000089510.5
Cenpb
centromere protein B
chr10_+_87982916
Show fit
0.44
ENSMUST00000169309.3
Nup37
nucleoporin 37
chr7_+_133239414
Show fit
0.44
ENSMUST00000128901.9
Edrf1
erythroid differentiation regulatory factor 1
chr8_-_26330783
Show fit
0.43
ENSMUST00000110608.3
ENSMUST00000068892.15
Ash2l
ASH2 like histone lysine methyltransferase complex subunit
chr9_+_25163735
Show fit
0.43
ENSMUST00000115272.9
ENSMUST00000165594.4
Septin7
septin 7
chr10_+_87982854
Show fit
0.42
ENSMUST00000052355.15
Nup37
nucleoporin 37
chr6_+_91492988
Show fit
0.42
ENSMUST00000206947.2
Lsm3
LSM3 homolog, U6 small nuclear RNA and mRNA degradation associated
chr7_-_80550968
Show fit
0.42
ENSMUST00000146402.2
ENSMUST00000026816.15
Wdr73
WD repeat domain 73
chr12_+_111504450
Show fit
0.41
ENSMUST00000166123.9
ENSMUST00000222441.2
Eif5
eukaryotic translation initiation factor 5
chr11_-_59730654
Show fit
0.41
ENSMUST00000019517.10
Cops3
COP9 signalosome subunit 3
chrX_+_161543384
Show fit
0.40
ENSMUST00000033720.12
ENSMUST00000112327.8
Rbbp7
retinoblastoma binding protein 7, chromatin remodeling factor
chr11_-_53191208
Show fit
0.40
ENSMUST00000020630.8
Hspa4
heat shock protein 4
chr17_-_34250616
Show fit
0.40
ENSMUST00000169397.9
Slc39a7
solute carrier family 39 (zinc transporter), member 7
chrX_+_108138965
Show fit
0.40
ENSMUST00000033598.9
Sh3bgrl
SH3-binding domain glutamic acid-rich protein like
chr10_-_67120959
Show fit
0.39
ENSMUST00000159002.2
ENSMUST00000077839.13
Nrbf2
nuclear receptor binding factor 2
chr13_+_55253102
Show fit
0.39
ENSMUST00000161315.8
ENSMUST00000021937.12
ENSMUST00000159278.8
ENSMUST00000159147.8
ENSMUST00000161077.2
Zfp346
zinc finger protein 346
chr9_+_99457829
Show fit
0.39
ENSMUST00000066650.12
ENSMUST00000148987.8
Dbr1
debranching RNA lariats 1
chr11_+_29122890
Show fit
0.39
ENSMUST00000102856.9
ENSMUST00000020755.12
Ppp4r3b
protein phosphatase 4 regulatory subunit 3B
chr15_-_31601932
Show fit
0.39
ENSMUST00000022842.16
Cct5
chaperonin containing Tcp1, subunit 5 (epsilon)
chr16_-_87292711
Show fit
0.39
ENSMUST00000176041.8
ENSMUST00000026704.14
Cct8
chaperonin containing Tcp1, subunit 8 (theta)
chr5_+_129864044
Show fit
0.39
ENSMUST00000201414.5
Cct6a
chaperonin containing Tcp1, subunit 6a (zeta)
chrX_+_161543423
Show fit
0.39
ENSMUST00000112326.8
Rbbp7
retinoblastoma binding protein 7, chromatin remodeling factor
chr17_+_29487881
Show fit
0.38
ENSMUST00000234845.2
ENSMUST00000235038.2
ENSMUST00000235050.2
ENSMUST00000120346.9
ENSMUST00000234377.2
ENSMUST00000235074.2
ENSMUST00000235040.2
ENSMUST00000234256.2
ENSMUST00000234459.2
BC004004
cDNA sequence BC004004
chr18_+_67523734
Show fit
0.38
ENSMUST00000001513.8
Tubb6
tubulin, beta 6 class V
chr15_-_98661076
Show fit
0.38
ENSMUST00000053183.12
Arf3
ADP-ribosylation factor 3
chr4_-_86775602
Show fit
0.37
ENSMUST00000102814.5
Rps6
ribosomal protein S6
chr6_+_91492910
Show fit
0.37
ENSMUST00000040607.6
Lsm3
LSM3 homolog, U6 small nuclear RNA and mRNA degradation associated
chr2_-_144112444
Show fit
0.36
ENSMUST00000028909.5
Snx5
sorting nexin 5
chr8_-_85696040
Show fit
0.36
ENSMUST00000214133.2
ENSMUST00000147812.8
Gm49661
Rnaseh2a
predicted gene, 49661
chr7_+_81412673
Show fit
0.36
ENSMUST00000042166.11
Ramac
RNA guanine-7 methyltransferase activating subunit
chr2_-_124965537
Show fit
0.36
ENSMUST00000142718.8
ENSMUST00000152367.8
ENSMUST00000067780.10
ENSMUST00000147105.8
Myef2
myelin basic protein expression factor 2, repressor
chr13_+_58550499
Show fit
0.36
ENSMUST00000225815.2
Rmi1
RecQ mediated genome instability 1
chr8_+_126396557
Show fit
0.36
ENSMUST00000053078.5
Map10
microtubule-associated protein 10
chr4_-_136776006
Show fit
0.36
ENSMUST00000049583.8
Zbtb40
zinc finger and BTB domain containing 40
chr14_+_31881822
Show fit
0.35
ENSMUST00000163336.8
ENSMUST00000169722.8
ENSMUST00000168385.8
Ncoa4
nuclear receptor coactivator 4
chr11_-_69470139
Show fit
0.35
ENSMUST00000048139.12
Wrap53
WD repeat containing, antisense to Trp53
chr12_+_8971603
Show fit
0.35
ENSMUST00000020909.4
Laptm4a
lysosomal-associated protein transmembrane 4A
chrX_-_7940959
Show fit
0.35
ENSMUST00000115636.4
ENSMUST00000115638.10
Suv39h1
suppressor of variegation 3-9 1
chr15_-_81756076
Show fit
0.35
ENSMUST00000023117.10
Phf5a
PHD finger protein 5A
chr8_+_85696216
Show fit
0.34
ENSMUST00000109734.8
ENSMUST00000005292.15
Prdx2
peroxiredoxin 2
chr2_+_36120438
Show fit
0.33
ENSMUST00000062069.6
Ptgs1
prostaglandin-endoperoxide synthase 1
chr5_+_30824121
Show fit
0.33
ENSMUST00000144742.6
ENSMUST00000149759.2
ENSMUST00000199320.5
Cenpa
centromere protein A
chr10_+_80634736
Show fit
0.33
ENSMUST00000147440.2
Sf3a2
splicing factor 3a, subunit 2
chr3_-_88317601
Show fit
0.33
ENSMUST00000193338.6
ENSMUST00000056370.13
Pmf1
polyamine-modulated factor 1
chr5_+_52991351
Show fit
0.33
ENSMUST00000031072.14
Anapc4
anaphase promoting complex subunit 4
chr17_+_25105617
Show fit
0.33
ENSMUST00000117890.8
ENSMUST00000168265.8
ENSMUST00000120943.8
ENSMUST00000068508.13
ENSMUST00000119829.8
Spsb3
splA/ryanodine receptor domain and SOCS box containing 3
chr16_+_48814548
Show fit
0.32
ENSMUST00000117994.8
ENSMUST00000048374.6
Cip2a
cell proliferation regulating inhibitor of protein phosphatase 2A
chr11_-_84761472
Show fit
0.32
ENSMUST00000018547.9
Ggnbp2
gametogenetin binding protein 2
chr7_-_117715394
Show fit
0.32
ENSMUST00000131374.8
Rps15a
ribosomal protein S15A
chr6_-_126916487
Show fit
0.32
ENSMUST00000144954.5
ENSMUST00000112220.8
ENSMUST00000112221.8
Rad51ap1
RAD51 associated protein 1
chr17_+_28547533
Show fit
0.32
ENSMUST00000233427.2
ENSMUST00000233937.2
Rpl10a
ribosomal protein L10A
chr1_-_80318028
Show fit
0.32
ENSMUST00000164108.8
Cul3
cullin 3
chr9_-_62029877
Show fit
0.32
ENSMUST00000185675.7
Glce
glucuronyl C5-epimerase
chr8_+_85696453
Show fit
0.31
ENSMUST00000125893.8
Prdx2
peroxiredoxin 2
chr11_-_95966407
Show fit
0.31
ENSMUST00000107686.8
ENSMUST00000107684.2
Atp5g1
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9)
chr3_+_137570334
Show fit
0.31
ENSMUST00000174561.8
ENSMUST00000173790.8
H2az1
H2A.Z variant histone 1
chr17_-_25105277
Show fit
0.31
ENSMUST00000234583.2
ENSMUST00000234968.2
ENSMUST00000044252.7
Nubp2
nucleotide binding protein 2
chr9_-_62719208
Show fit
0.31
ENSMUST00000034775.10
Fem1b
fem 1 homolog b
chr2_+_60040231
Show fit
0.31
ENSMUST00000102748.11
Marchf7
membrane associated ring-CH-type finger 7
chr16_-_22258469
Show fit
0.31
ENSMUST00000079601.13
Etv5
ets variant 5
chr8_+_95564949
Show fit
0.31
ENSMUST00000034234.15
ENSMUST00000159871.4
Coq9
coenzyme Q9
chr17_+_21709244
Show fit
0.30
ENSMUST00000233158.2
Zfp53
zinc finger protein 53
chr17_+_56347424
Show fit
0.30
ENSMUST00000002914.10
Chaf1a
chromatin assembly factor 1, subunit A (p150)
chr15_+_76227695
Show fit
0.30
ENSMUST00000023210.8
ENSMUST00000231045.2
Cyc1
cytochrome c-1
chr2_-_168048795
Show fit
0.29
ENSMUST00000057793.11
Adnp
activity-dependent neuroprotective protein
chr13_+_59733163
Show fit
0.29
ENSMUST00000166923.9
Naa35
N(alpha)-acetyltransferase 35, NatC auxiliary subunit
chr13_-_58550290
Show fit
0.29
ENSMUST00000043269.14
ENSMUST00000177060.8
ENSMUST00000224182.2
ENSMUST00000176207.8
Hnrnpk
heterogeneous nuclear ribonucleoprotein K
chr4_-_120782198
Show fit
0.29
ENSMUST00000030375.9
ENSMUST00000177880.2
ENSMUST00000156836.2
Exo5
exonuclease 5
chr11_-_53321242
Show fit
0.29
ENSMUST00000109019.8
Uqcrq
ubiquinol-cytochrome c reductase, complex III subunit VII
chr10_+_11219117
Show fit
0.29
ENSMUST00000069106.5
Epm2a
epilepsy, progressive myoclonic epilepsy, type 2 gene alpha
chr10_+_127102193
Show fit
0.29
ENSMUST00000026479.11
Dctn2
dynactin 2
chr2_+_32851571
Show fit
0.29
ENSMUST00000126610.2
Rpl12
ribosomal protein L12
chr11_-_33113071
Show fit
0.28
ENSMUST00000093201.13
ENSMUST00000101375.5
ENSMUST00000109354.10
ENSMUST00000075641.10
Npm1
nucleophosmin 1
chr11_-_107080150
Show fit
0.28
ENSMUST00000106757.8
ENSMUST00000018577.8
Nol11
nucleolar protein 11
chr11_+_74788904
Show fit
0.27
ENSMUST00000045807.14
Tsr1
TSR1 20S rRNA accumulation
chr12_+_4132567
Show fit
0.27
ENSMUST00000020986.15
ENSMUST00000049584.6
Dnajc27
DnaJ heat shock protein family (Hsp40) member C27
chr14_+_58063668
Show fit
0.27
ENSMUST00000022538.5
Mrpl57
mitochondrial ribosomal protein L57
chr6_+_83303052
Show fit
0.27
ENSMUST00000038658.15
Mob1a
MOB kinase activator 1A
chr3_-_107538996
Show fit
0.27
ENSMUST00000064759.7
Strip1
striatin interacting protein 1
chr7_+_44117511
Show fit
0.27
ENSMUST00000121922.3
ENSMUST00000208117.2
Josd2
Josephin domain containing 2
chr13_-_38178059
Show fit
0.26
ENSMUST00000225319.2
ENSMUST00000225246.2
ENSMUST00000021864.8
Ssr1
signal sequence receptor, alpha
chr3_-_86049988
Show fit
0.26
ENSMUST00000029722.7
Rps3a1
ribosomal protein S3A1
chr7_-_44773750
Show fit
0.26
ENSMUST00000211725.2
ENSMUST00000003521.10
Rps11
ribosomal protein S11
chr11_-_84761538
Show fit
0.26
ENSMUST00000170741.2
ENSMUST00000172405.8
ENSMUST00000100686.10
ENSMUST00000108081.9
Ggnbp2
gametogenetin binding protein 2
chr4_+_94502719
Show fit
0.26
ENSMUST00000107104.3
ENSMUST00000030311.11
Ift74
intraflagellar transport 74
chr19_+_21630887
Show fit
0.26
ENSMUST00000052556.5
Abhd17b
abhydrolase domain containing 17B
chr11_-_120524362
Show fit
0.26
ENSMUST00000058162.14
Mafg
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian)
chr16_-_16942970
Show fit
0.26
ENSMUST00000093336.8
ENSMUST00000231681.2
2610318N02Rik
RIKEN cDNA 2610318N02 gene
chr9_+_35179153
Show fit
0.25
ENSMUST00000034543.5
Rpusd4
RNA pseudouridylate synthase domain containing 4
chr8_-_26244279
Show fit
0.25
ENSMUST00000033975.9
Ddhd2
DDHD domain containing 2
chr18_+_35695199
Show fit
0.25
ENSMUST00000236860.2
ENSMUST00000166793.10
ENSMUST00000237780.2
ENSMUST00000236507.2
ENSMUST00000235960.2
ENSMUST00000237061.2
Matr3
matrin 3
chr7_-_28981335
Show fit
0.25
ENSMUST00000108236.5
ENSMUST00000098604.12
Spint2
serine protease inhibitor, Kunitz type 2
chr15_-_97603664
Show fit
0.25
ENSMUST00000023104.7
Rpap3
RNA polymerase II associated protein 3
chr4_-_134262694
Show fit
0.25
ENSMUST00000116279.10
ENSMUST00000146808.2
Mtfr1l
mitochondrial fission regulator 1-like
chr6_-_91492860
Show fit
0.25
ENSMUST00000206476.2
ENSMUST00000032182.5
Xpc
xeroderma pigmentosum, complementation group C
chr4_-_133972890
Show fit
0.25
ENSMUST00000030644.8
Zfp593
zinc finger protein 593
chr2_+_26800757
Show fit
0.24
ENSMUST00000102898.5
Rpl7a
ribosomal protein L7A
chr6_+_133082202
Show fit
0.24
ENSMUST00000191462.2
Smim10l1
small integral membrane protein 10 like 1
chr12_+_111504640
Show fit
0.24
ENSMUST00000222375.2
ENSMUST00000222388.2
Eif5
eukaryotic translation initiation factor 5
chr7_+_44117444
Show fit
0.24
ENSMUST00000206887.2
ENSMUST00000117324.8
ENSMUST00000120852.8
ENSMUST00000134398.3
ENSMUST00000118628.8
Josd2
Josephin domain containing 2
chr12_+_108758871
Show fit
0.24
ENSMUST00000021692.9
Yy1
YY1 transcription factor
chr15_-_83033508
Show fit
0.24
ENSMUST00000100375.11
Poldip3
polymerase (DNA-directed), delta interacting protein 3
chr7_+_98352298
Show fit
0.24
ENSMUST00000033009.16
Thap12
THAP domain containing 12
chr13_-_58549917
Show fit
0.24
ENSMUST00000176305.8
ENSMUST00000176558.2
ENSMUST00000225674.2
ENSMUST00000176849.8
Hnrnpk
heterogeneous nuclear ribonucleoprotein K
chr19_+_38919353
Show fit
0.24
ENSMUST00000025965.12
Hells
helicase, lymphoid specific
chr7_-_46360066
Show fit
0.24
ENSMUST00000143082.4
Saal1
serum amyloid A-like 1
chr8_+_85696396
Show fit
0.24
ENSMUST00000109733.8
Prdx2
peroxiredoxin 2
chr3_-_68911886
Show fit
0.23
ENSMUST00000169064.8
Ift80
intraflagellar transport 80
chr9_+_107828136
Show fit
0.23
ENSMUST00000049348.9
ENSMUST00000194271.2
Traip
TRAF-interacting protein
chr9_+_58489523
Show fit
0.23
ENSMUST00000177292.8
ENSMUST00000085651.12
ENSMUST00000176557.8
ENSMUST00000114121.11
ENSMUST00000177064.8
Nptn
neuroplastin