Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
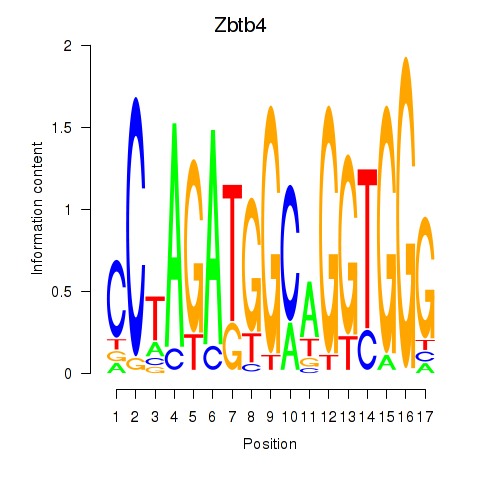
Results for Zbtb4
Z-value: 1.49
Transcription factors associated with Zbtb4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb4
|
ENSMUSG00000018750.15 | zinc finger and BTB domain containing 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb4 | mm39_v1_chr11_+_69656725_69656752 | -0.96 | 8.4e-03 | Click! |
Activity profile of Zbtb4 motif
Sorted Z-values of Zbtb4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_141028539 | 1.31 |
ENSMUST00000006614.3
|
Epha2
|
Eph receptor A2 |
| chr8_-_78451055 | 1.15 |
ENSMUST00000034029.8
|
Ednra
|
endothelin receptor type A |
| chr4_-_156340276 | 0.86 |
ENSMUST00000220228.2
ENSMUST00000218788.2 ENSMUST00000179919.3 |
Samd11
|
sterile alpha motif domain containing 11 |
| chr17_-_34406193 | 0.84 |
ENSMUST00000173831.3
|
Psmb9
|
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) |
| chr4_+_102617495 | 0.83 |
ENSMUST00000072481.12
ENSMUST00000156596.8 ENSMUST00000080728.13 ENSMUST00000106882.9 |
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr15_+_100659622 | 0.82 |
ENSMUST00000023776.13
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr10_+_80765900 | 0.81 |
ENSMUST00000015456.10
ENSMUST00000220246.2 |
Gadd45b
|
growth arrest and DNA-damage-inducible 45 beta |
| chr11_-_69786324 | 0.75 |
ENSMUST00000001631.7
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr4_+_6365650 | 0.74 |
ENSMUST00000029912.11
ENSMUST00000103008.12 |
Sdcbp
|
syndecan binding protein |
| chr4_-_135699205 | 0.74 |
ENSMUST00000105852.8
|
Lypla2
|
lysophospholipase 2 |
| chr4_+_102617332 | 0.73 |
ENSMUST00000066824.14
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr4_+_6365694 | 0.73 |
ENSMUST00000175769.8
ENSMUST00000140830.8 ENSMUST00000108374.8 |
Sdcbp
|
syndecan binding protein |
| chr8_+_39472981 | 0.71 |
ENSMUST00000239508.1
ENSMUST00000239509.1 |
TUSC3
|
tumor suppressor candidate 3 |
| chr1_+_39406979 | 0.69 |
ENSMUST00000178079.8
ENSMUST00000179954.8 |
Rpl31
|
ribosomal protein L31 |
| chr16_-_22946441 | 0.69 |
ENSMUST00000133847.9
ENSMUST00000115338.8 ENSMUST00000023598.15 |
Rfc4
|
replication factor C (activator 1) 4 |
| chr5_-_21087023 | 0.65 |
ENSMUST00000118174.8
|
Phtf2
|
putative homeodomain transcription factor 2 |
| chr11_-_22810467 | 0.63 |
ENSMUST00000055549.4
|
B3gnt2
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 |
| chr9_+_69361348 | 0.63 |
ENSMUST00000134907.8
|
Anxa2
|
annexin A2 |
| chr10_-_35587888 | 0.62 |
ENSMUST00000080898.4
|
Amd2
|
S-adenosylmethionine decarboxylase 2 |
| chr12_-_54703281 | 0.59 |
ENSMUST00000056228.8
|
Sptssa
|
serine palmitoyltransferase, small subunit A |
| chr8_+_106785434 | 0.56 |
ENSMUST00000212742.2
ENSMUST00000211991.2 |
Nfatc3
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
| chr5_-_121025645 | 0.54 |
ENSMUST00000086368.12
|
Oas1g
|
2'-5' oligoadenylate synthetase 1G |
| chr11_-_6469494 | 0.52 |
ENSMUST00000134489.2
|
Myo1g
|
myosin IG |
| chr14_+_121148625 | 0.51 |
ENSMUST00000032898.9
|
Ipo5
|
importin 5 |
| chr17_-_74601769 | 0.51 |
ENSMUST00000078459.8
ENSMUST00000232989.2 |
Memo1
|
mediator of cell motility 1 |
| chr17_+_34406762 | 0.49 |
ENSMUST00000041633.15
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr7_-_141119703 | 0.49 |
ENSMUST00000118694.8
ENSMUST00000153191.8 ENSMUST00000026586.13 ENSMUST00000166082.8 |
Chid1
|
chitinase domain containing 1 |
| chr10_-_80426125 | 0.46 |
ENSMUST00000187646.2
ENSMUST00000191440.7 ENSMUST00000003436.12 |
Abhd17a
|
abhydrolase domain containing 17A |
| chr17_+_34406523 | 0.45 |
ENSMUST00000170086.8
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr7_-_125968653 | 0.44 |
ENSMUST00000205642.2
ENSMUST00000032997.8 ENSMUST00000206793.2 |
Lat
|
linker for activation of T cells |
| chr6_-_126916487 | 0.43 |
ENSMUST00000144954.5
ENSMUST00000112220.8 ENSMUST00000112221.8 |
Rad51ap1
|
RAD51 associated protein 1 |
| chr7_+_126446588 | 0.43 |
ENSMUST00000141805.8
ENSMUST00000064110.14 ENSMUST00000205938.2 ENSMUST00000152051.8 |
Doc2a
|
double C2, alpha |
| chr17_-_37269153 | 0.43 |
ENSMUST00000172823.2
|
Polr1h
|
RNA polymerase I subunit H |
| chr5_+_36622342 | 0.42 |
ENSMUST00000031099.4
|
Grpel1
|
GrpE-like 1, mitochondrial |
| chr3_-_90603013 | 0.42 |
ENSMUST00000069960.12
ENSMUST00000117167.2 |
S100a9
|
S100 calcium binding protein A9 (calgranulin B) |
| chr14_+_67953547 | 0.42 |
ENSMUST00000078053.13
|
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr2_-_118987305 | 0.41 |
ENSMUST00000135419.8
ENSMUST00000129351.2 ENSMUST00000139519.2 ENSMUST00000094695.12 |
Rmdn3
|
regulator of microtubule dynamics 3 |
| chr19_+_46695889 | 0.41 |
ENSMUST00000003655.9
|
As3mt
|
arsenite methyltransferase |
| chr1_+_127796508 | 0.40 |
ENSMUST00000037649.6
ENSMUST00000212506.2 |
Rab3gap1
|
RAB3 GTPase activating protein subunit 1 |
| chr14_-_30723549 | 0.40 |
ENSMUST00000226782.2
ENSMUST00000186131.7 ENSMUST00000228767.2 |
Spcs1
|
signal peptidase complex subunit 1 homolog (S. cerevisiae) |
| chr17_-_37269330 | 0.40 |
ENSMUST00000113669.9
|
Polr1h
|
RNA polymerase I subunit H |
| chr7_-_139696308 | 0.39 |
ENSMUST00000026538.13
|
Echs1
|
enoyl Coenzyme A hydratase, short chain, 1, mitochondrial |
| chr5_+_145020910 | 0.39 |
ENSMUST00000124379.3
|
Arpc1a
|
actin related protein 2/3 complex, subunit 1A |
| chr16_+_32249713 | 0.39 |
ENSMUST00000115137.8
ENSMUST00000079791.11 |
Pcyt1a
|
phosphate cytidylyltransferase 1, choline, alpha isoform |
| chr7_-_34089109 | 0.38 |
ENSMUST00000085585.12
|
Lsm14a
|
LSM14A mRNA processing body assembly factor |
| chr10_-_75617245 | 0.38 |
ENSMUST00000001715.10
|
Gstt3
|
glutathione S-transferase, theta 3 |
| chr2_-_144112444 | 0.38 |
ENSMUST00000028909.5
|
Snx5
|
sorting nexin 5 |
| chr6_+_145692439 | 0.37 |
ENSMUST00000111704.8
|
Rassf8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr11_-_51579441 | 0.37 |
ENSMUST00000007921.9
|
0610009B22Rik
|
RIKEN cDNA 0610009B22 gene |
| chr2_-_134486039 | 0.36 |
ENSMUST00000038228.11
|
Tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr1_-_180641430 | 0.35 |
ENSMUST00000162814.8
|
H3f3a
|
H3.3 histone A |
| chr5_+_145020640 | 0.35 |
ENSMUST00000031625.15
|
Arpc1a
|
actin related protein 2/3 complex, subunit 1A |
| chr12_+_111505253 | 0.34 |
ENSMUST00000220803.2
|
Eif5
|
eukaryotic translation initiation factor 5 |
| chr4_+_119052693 | 0.34 |
ENSMUST00000097908.4
|
Svbp
|
small vasohibin binding protein |
| chr8_-_23083829 | 0.34 |
ENSMUST00000179233.2
|
Vdac3
|
voltage-dependent anion channel 3 |
| chr6_+_58808733 | 0.33 |
ENSMUST00000126292.8
ENSMUST00000031823.12 |
Herc3
|
hect domain and RLD 3 |
| chr11_-_69728560 | 0.33 |
ENSMUST00000108634.9
|
Nlgn2
|
neuroligin 2 |
| chr5_+_21087107 | 0.33 |
ENSMUST00000115259.3
|
Tmem60
|
transmembrane protein 60 |
| chr17_+_56316305 | 0.32 |
ENSMUST00000159340.8
|
Mpnd
|
MPN domain containing |
| chr17_+_8502682 | 0.32 |
ENSMUST00000124023.8
|
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr13_-_63036096 | 0.31 |
ENSMUST00000092888.11
|
Fbp1
|
fructose bisphosphatase 1 |
| chr15_+_73595012 | 0.31 |
ENSMUST00000230044.2
|
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr2_+_148522943 | 0.30 |
ENSMUST00000028928.8
|
Gzf1
|
GDNF-inducible zinc finger protein 1 |
| chr17_-_37269425 | 0.29 |
ENSMUST00000172518.8
|
Polr1h
|
RNA polymerase I subunit H |
| chr2_-_26406631 | 0.29 |
ENSMUST00000132820.2
|
Notch1
|
notch 1 |
| chr14_+_30856687 | 0.29 |
ENSMUST00000090212.5
|
Nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr5_-_110434026 | 0.29 |
ENSMUST00000031472.12
|
Pxmp2
|
peroxisomal membrane protein 2 |
| chrX_+_93768175 | 0.29 |
ENSMUST00000101388.4
|
Zxdb
|
zinc finger, X-linked, duplicated B |
| chr9_+_106080307 | 0.29 |
ENSMUST00000024047.12
ENSMUST00000216348.2 |
Twf2
|
twinfilin actin binding protein 2 |
| chr2_-_71198091 | 0.29 |
ENSMUST00000151937.8
|
Slc25a12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
| chr17_-_29483075 | 0.28 |
ENSMUST00000024802.10
|
Ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr4_+_130253925 | 0.28 |
ENSMUST00000105994.4
|
Snrnp40
|
small nuclear ribonucleoprotein 40 (U5) |
| chr16_+_56942050 | 0.28 |
ENSMUST00000166897.3
|
Tomm70a
|
translocase of outer mitochondrial membrane 70A |
| chr3_+_65435825 | 0.28 |
ENSMUST00000047906.10
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr17_+_56316201 | 0.27 |
ENSMUST00000149441.8
ENSMUST00000162883.8 ENSMUST00000159996.8 |
Mpnd
|
MPN domain containing |
| chr14_-_30723292 | 0.27 |
ENSMUST00000228736.2
ENSMUST00000226374.2 |
Spcs1
|
signal peptidase complex subunit 1 homolog (S. cerevisiae) |
| chr18_-_35064906 | 0.27 |
ENSMUST00000025218.8
|
Etf1
|
eukaryotic translation termination factor 1 |
| chr14_+_67953687 | 0.27 |
ENSMUST00000150768.8
|
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr15_-_35938155 | 0.27 |
ENSMUST00000156915.3
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chr14_-_54651442 | 0.27 |
ENSMUST00000227334.2
|
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr3_+_60910207 | 0.26 |
ENSMUST00000029331.7
ENSMUST00000193201.2 ENSMUST00000193943.2 |
P2ry1
|
purinergic receptor P2Y, G-protein coupled 1 |
| chr9_+_69902697 | 0.26 |
ENSMUST00000165389.8
|
Bnip2
|
BCL2/adenovirus E1B interacting protein 2 |
| chr17_-_26161797 | 0.25 |
ENSMUST00000208043.2
ENSMUST00000148382.2 ENSMUST00000145745.3 |
Pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr9_+_15404728 | 0.25 |
ENSMUST00000215907.2
|
Smco4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr9_+_60701749 | 0.24 |
ENSMUST00000214354.2
ENSMUST00000050183.7 ENSMUST00000217656.2 |
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr17_+_8502594 | 0.24 |
ENSMUST00000155364.8
ENSMUST00000046754.15 |
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chrX_-_92875712 | 0.24 |
ENSMUST00000045748.7
|
Pdk3
|
pyruvate dehydrogenase kinase, isoenzyme 3 |
| chr2_+_30306116 | 0.24 |
ENSMUST00000113601.10
ENSMUST00000113603.10 |
Ptpa
|
protein phosphatase 2 protein activator |
| chr2_+_25313240 | 0.23 |
ENSMUST00000134259.8
ENSMUST00000100320.5 |
Fut7
|
fucosyltransferase 7 |
| chr1_-_93270430 | 0.23 |
ENSMUST00000027493.4
|
Pask
|
PAS domain containing serine/threonine kinase |
| chr11_-_78427061 | 0.23 |
ENSMUST00000017759.9
ENSMUST00000108277.3 |
Tnfaip1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr2_-_26917921 | 0.23 |
ENSMUST00000102890.11
ENSMUST00000153388.2 ENSMUST00000045702.6 |
Slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr11_-_69470139 | 0.23 |
ENSMUST00000048139.12
|
Wrap53
|
WD repeat containing, antisense to Trp53 |
| chr5_-_123126550 | 0.23 |
ENSMUST00000086200.11
ENSMUST00000156474.8 |
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr13_+_85337552 | 0.22 |
ENSMUST00000165077.8
ENSMUST00000164127.8 ENSMUST00000163600.8 |
Ccnh
|
cyclin H |
| chr7_+_19115929 | 0.22 |
ENSMUST00000062831.16
ENSMUST00000108461.8 ENSMUST00000108460.8 |
Ercc2
|
excision repair cross-complementing rodent repair deficiency, complementation group 2 |
| chr5_+_24633206 | 0.22 |
ENSMUST00000115049.9
|
Slc4a2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr14_+_47069582 | 0.21 |
ENSMUST00000068532.10
|
Cgrrf1
|
cell growth regulator with ring finger domain 1 |
| chr9_+_106158549 | 0.21 |
ENSMUST00000191434.2
|
Poc1a
|
POC1 centriolar protein A |
| chrX_-_55643429 | 0.21 |
ENSMUST00000059899.3
|
Mmgt1
|
membrane magnesium transporter 1 |
| chr17_-_74601828 | 0.21 |
ENSMUST00000233514.3
|
Memo1
|
mediator of cell motility 1 |
| chr17_+_26780486 | 0.20 |
ENSMUST00000001619.16
|
Ergic1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
| chr9_+_75221415 | 0.20 |
ENSMUST00000215875.2
|
Gnb5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chr7_-_34088886 | 0.20 |
ENSMUST00000155256.8
|
Lsm14a
|
LSM14A mRNA processing body assembly factor |
| chr4_+_156194427 | 0.20 |
ENSMUST00000072554.13
ENSMUST00000169550.8 ENSMUST00000105576.2 |
9430015G10Rik
|
RIKEN cDNA 9430015G10 gene |
| chr19_-_46384158 | 0.20 |
ENSMUST00000237742.2
ENSMUST00000040270.6 |
Actr1a
|
ARP1 actin-related protein 1A, centractin alpha |
| chr4_+_124960465 | 0.20 |
ENSMUST00000052183.7
|
Snip1
|
Smad nuclear interacting protein 1 |
| chr17_-_34243608 | 0.20 |
ENSMUST00000025183.9
|
Ring1
|
ring finger protein 1 |
| chr7_+_16472335 | 0.20 |
ENSMUST00000086112.8
ENSMUST00000205607.2 |
Ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr15_+_76215488 | 0.19 |
ENSMUST00000172281.8
|
Gpaa1
|
GPI anchor attachment protein 1 |
| chr10_+_78261503 | 0.19 |
ENSMUST00000005185.8
|
Cstb
|
cystatin B |
| chr4_+_119052548 | 0.19 |
ENSMUST00000106345.3
|
Svbp
|
small vasohibin binding protein |
| chr2_-_84500951 | 0.19 |
ENSMUST00000189988.3
ENSMUST00000189636.8 ENSMUST00000102646.4 ENSMUST00000102647.11 ENSMUST00000117299.10 |
Selenoh
|
selenoprotein H |
| chr4_+_10874498 | 0.19 |
ENSMUST00000080517.14
|
2610301B20Rik
|
RIKEN cDNA 2610301B20 gene |
| chr19_-_29790352 | 0.19 |
ENSMUST00000099525.5
|
Ranbp6
|
RAN binding protein 6 |
| chr4_+_129823042 | 0.19 |
ENSMUST00000084263.6
|
Spocd1
|
SPOC domain containing 1 |
| chr15_-_58695379 | 0.19 |
ENSMUST00000072113.6
|
Tmem65
|
transmembrane protein 65 |
| chr12_+_111505216 | 0.19 |
ENSMUST00000050993.11
|
Eif5
|
eukaryotic translation initiation factor 5 |
| chr13_-_65260716 | 0.18 |
ENSMUST00000054730.10
ENSMUST00000155487.8 |
Mfsd14b
|
major facilitator superfamily domain containing 14B |
| chr5_-_36905983 | 0.18 |
ENSMUST00000071949.5
|
Bloc1s4
|
biogenesis of lysosomal organelles complex-1, subunit 4, cappuccino |
| chr1_-_152262425 | 0.18 |
ENSMUST00000015124.15
|
Tsen15
|
tRNA splicing endonuclease subunit 15 |
| chr14_+_65612788 | 0.18 |
ENSMUST00000224687.2
|
Zfp395
|
zinc finger protein 395 |
| chr1_-_184543367 | 0.18 |
ENSMUST00000048462.13
ENSMUST00000110992.9 |
Mtarc1
|
mitochondrial amidoxime reducing component 1 |
| chr2_-_150746574 | 0.17 |
ENSMUST00000056149.15
|
Abhd12
|
abhydrolase domain containing 12 |
| chr7_+_80510658 | 0.17 |
ENSMUST00000132163.8
ENSMUST00000205361.2 ENSMUST00000147125.2 |
Zscan2
|
zinc finger and SCAN domain containing 2 |
| chr15_+_76235503 | 0.17 |
ENSMUST00000023212.15
ENSMUST00000160172.8 |
Maf1
|
MAF1 homolog, negative regulator of RNA polymerase III |
| chr2_+_128659997 | 0.17 |
ENSMUST00000110325.8
|
Tmem87b
|
transmembrane protein 87B |
| chr4_-_132649798 | 0.17 |
ENSMUST00000097856.10
ENSMUST00000030696.11 |
Fam76a
|
family with sequence similarity 76, member A |
| chr2_+_84891281 | 0.16 |
ENSMUST00000238769.2
|
Tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr2_+_179899166 | 0.16 |
ENSMUST00000059080.7
|
Rps21
|
ribosomal protein S21 |
| chr2_+_128660212 | 0.16 |
ENSMUST00000143398.3
ENSMUST00000152210.2 |
Tmem87b
|
transmembrane protein 87B |
| chr13_+_12580743 | 0.15 |
ENSMUST00000221560.2
ENSMUST00000071973.8 |
Ero1b
|
endoplasmic reticulum oxidoreductase 1 beta |
| chr6_+_48818567 | 0.15 |
ENSMUST00000203639.3
|
Tmem176a
|
transmembrane protein 176A |
| chr9_+_37400317 | 0.15 |
ENSMUST00000239459.2
|
Msantd2
|
Myb/SANT-like DNA-binding domain containing 2 |
| chr6_+_58808438 | 0.15 |
ENSMUST00000141600.8
ENSMUST00000122981.2 |
Herc3
|
hect domain and RLD 3 |
| chr15_-_43146092 | 0.15 |
ENSMUST00000022960.4
|
Eif3e
|
eukaryotic translation initiation factor 3, subunit E |
| chr4_-_130253703 | 0.15 |
ENSMUST00000134159.3
|
Zcchc17
|
zinc finger, CCHC domain containing 17 |
| chr2_+_60040231 | 0.14 |
ENSMUST00000102748.11
|
Marchf7
|
membrane associated ring-CH-type finger 7 |
| chr1_-_152262339 | 0.14 |
ENSMUST00000162371.2
|
Tsen15
|
tRNA splicing endonuclease subunit 15 |
| chrX_+_151922936 | 0.14 |
ENSMUST00000039720.11
ENSMUST00000144175.3 |
Rragb
|
Ras-related GTP binding B |
| chr4_+_150938376 | 0.14 |
ENSMUST00000073600.9
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr8_-_120828211 | 0.14 |
ENSMUST00000034280.9
|
Zdhhc7
|
zinc finger, DHHC domain containing 7 |
| chr2_+_30306045 | 0.14 |
ENSMUST00000042055.10
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr1_+_39406954 | 0.14 |
ENSMUST00000194746.6
|
Rpl31
|
ribosomal protein L31 |
| chr3_+_127583550 | 0.13 |
ENSMUST00000163775.6
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr7_-_84661476 | 0.13 |
ENSMUST00000124773.3
ENSMUST00000233725.2 ENSMUST00000233739.2 ENSMUST00000232837.2 |
Vmn2r66
|
vomeronasal 2, receptor 66 |
| chr6_-_48818430 | 0.13 |
ENSMUST00000205147.3
|
Tmem176b
|
transmembrane protein 176B |
| chr9_-_37166699 | 0.13 |
ENSMUST00000161114.2
|
Slc37a2
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 2 |
| chr6_-_91492860 | 0.13 |
ENSMUST00000206476.2
ENSMUST00000032182.5 |
Xpc
|
xeroderma pigmentosum, complementation group C |
| chr4_+_10874537 | 0.12 |
ENSMUST00000101504.3
|
2610301B20Rik
|
RIKEN cDNA 2610301B20 gene |
| chr9_+_21838767 | 0.12 |
ENSMUST00000006403.7
ENSMUST00000170304.9 ENSMUST00000216710.2 |
Ccdc159
|
coiled-coil domain containing 159 |
| chr11_+_50065666 | 0.12 |
ENSMUST00000020647.10
|
Mrnip
|
MRN complex interacting protein |
| chr11_-_3477916 | 0.12 |
ENSMUST00000020718.10
|
Smtn
|
smoothelin |
| chr18_+_11790409 | 0.12 |
ENSMUST00000047322.8
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chrX_+_102465616 | 0.12 |
ENSMUST00000182089.2
|
Gm26992
|
predicted gene, 26992 |
| chr1_-_165064455 | 0.12 |
ENSMUST00000043235.8
|
Tiprl
|
TIP41, TOR signalling pathway regulator-like (S. cerevisiae) |
| chr3_+_65435878 | 0.11 |
ENSMUST00000130705.2
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr17_+_6926452 | 0.11 |
ENSMUST00000097430.10
|
Sytl3
|
synaptotagmin-like 3 |
| chr9_-_22046970 | 0.11 |
ENSMUST00000165735.9
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr3_+_152101080 | 0.11 |
ENSMUST00000106103.8
|
Zzz3
|
zinc finger, ZZ domain containing 3 |
| chr7_+_28682253 | 0.10 |
ENSMUST00000085835.8
|
Map4k1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr14_+_67953584 | 0.10 |
ENSMUST00000145542.8
ENSMUST00000125212.2 |
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr11_-_59363289 | 0.10 |
ENSMUST00000057799.8
|
Zfp867
|
zinc finger protein 867 |
| chr15_-_89261242 | 0.10 |
ENSMUST00000023285.5
|
Tymp
|
thymidine phosphorylase |
| chr19_-_40600619 | 0.10 |
ENSMUST00000132452.2
ENSMUST00000135795.8 ENSMUST00000025981.15 |
Tctn3
|
tectonic family member 3 |
| chr19_-_11582207 | 0.10 |
ENSMUST00000025582.11
|
Ms4a6d
|
membrane-spanning 4-domains, subfamily A, member 6D |
| chr8_+_105065980 | 0.09 |
ENSMUST00000212139.2
|
Cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr9_+_96078340 | 0.09 |
ENSMUST00000034982.16
ENSMUST00000188008.7 ENSMUST00000188750.7 ENSMUST00000185644.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr11_+_121593219 | 0.09 |
ENSMUST00000036742.14
|
Metrnl
|
meteorin, glial cell differentiation regulator-like |
| chr17_+_26780453 | 0.09 |
ENSMUST00000167662.8
|
Ergic1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
| chr9_-_42383494 | 0.09 |
ENSMUST00000128959.8
ENSMUST00000066148.12 ENSMUST00000138506.8 |
Tbcel
|
tubulin folding cofactor E-like |
| chr16_+_32250043 | 0.08 |
ENSMUST00000115140.2
ENSMUST00000104893.10 |
Pcyt1a
|
phosphate cytidylyltransferase 1, choline, alpha isoform |
| chr5_-_34345014 | 0.08 |
ENSMUST00000042701.13
ENSMUST00000119171.2 |
Mxd4
|
Max dimerization protein 4 |
| chr16_+_36648728 | 0.08 |
ENSMUST00000114819.8
ENSMUST00000023535.4 |
Iqcb1
|
IQ calmodulin-binding motif containing 1 |
| chr7_+_87927293 | 0.08 |
ENSMUST00000032779.12
ENSMUST00000131108.9 ENSMUST00000128791.2 |
Ctsc
|
cathepsin C |
| chr6_-_57802131 | 0.07 |
ENSMUST00000204878.3
ENSMUST00000145608.7 ENSMUST00000203212.3 ENSMUST00000114297.5 |
Vopp1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr11_+_84070678 | 0.07 |
ENSMUST00000136463.9
|
Acaca
|
acetyl-Coenzyme A carboxylase alpha |
| chr9_+_108367801 | 0.07 |
ENSMUST00000006854.13
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr9_+_81745723 | 0.07 |
ENSMUST00000057067.10
ENSMUST00000189832.7 ENSMUST00000189391.2 |
Mei4
|
meiotic double-stranded break formation protein 4 |
| chr8_+_105991280 | 0.07 |
ENSMUST00000036221.12
|
Fbxl8
|
F-box and leucine-rich repeat protein 8 |
| chr5_+_77099229 | 0.07 |
ENSMUST00000141687.2
|
Paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoribosylaminoimidazole, succinocarboxamide synthetase |
| chr3_-_135373560 | 0.07 |
ENSMUST00000164430.7
|
Nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
| chr17_-_45997823 | 0.06 |
ENSMUST00000156254.8
|
Tmem63b
|
transmembrane protein 63b |
| chr17_+_29833760 | 0.06 |
ENSMUST00000024817.15
ENSMUST00000162588.3 |
Rnf8
|
ring finger protein 8 |
| chr6_-_49240944 | 0.06 |
ENSMUST00000204189.3
ENSMUST00000031841.9 |
Tra2a
|
transformer 2 alpha |
| chr11_-_53321606 | 0.06 |
ENSMUST00000061326.5
ENSMUST00000109021.4 |
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr10_-_80850712 | 0.06 |
ENSMUST00000126317.2
ENSMUST00000092285.10 ENSMUST00000117805.8 |
Gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr15_+_73594965 | 0.06 |
ENSMUST00000165541.8
ENSMUST00000167582.8 |
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr3_-_83749036 | 0.05 |
ENSMUST00000029623.11
|
Tlr2
|
toll-like receptor 2 |
| chr7_-_35346553 | 0.05 |
ENSMUST00000120714.2
|
Pdcd5
|
programmed cell death 5 |
| chr10_+_39608094 | 0.05 |
ENSMUST00000019986.13
|
Rev3l
|
REV3 like, DNA directed polymerase zeta catalytic subunit |
| chr9_-_106035308 | 0.05 |
ENSMUST00000159809.2
ENSMUST00000162562.2 ENSMUST00000036382.13 |
Glyctk
|
glycerate kinase |
| chr8_+_84682136 | 0.05 |
ENSMUST00000005607.9
|
Asf1b
|
anti-silencing function 1B histone chaperone |
| chr10_+_129223504 | 0.05 |
ENSMUST00000082342.3
|
Olfr784
|
olfactory receptor 784 |
| chr4_-_148244028 | 0.05 |
ENSMUST00000167160.8
ENSMUST00000151246.8 |
Fbxo44
|
F-box protein 44 |
| chr5_-_77099406 | 0.05 |
ENSMUST00000140076.2
|
Ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr17_-_46581697 | 0.05 |
ENSMUST00000224055.2
|
Tjap1
|
tight junction associated protein 1 |
| chr10_-_29238028 | 0.05 |
ENSMUST00000160372.9
ENSMUST00000214896.2 |
Rnf146
|
ring finger protein 146 |
| chr3_-_69506293 | 0.04 |
ENSMUST00000061826.3
|
B3galnt1
|
UDP-GalNAc:betaGlcNAc beta 1,3-galactosaminyltransferase, polypeptide 1 |
| chr11_-_62680228 | 0.04 |
ENSMUST00000207597.2
ENSMUST00000108705.8 |
Zfp286
|
zinc finger protein 286 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.3 | 0.9 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.2 | 1.5 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.2 | 0.6 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.2 | 0.6 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.2 | 0.6 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.4 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.4 | GO:1903233 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 0.1 | 0.5 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.1 | 0.4 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.1 | 0.7 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.3 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 0.6 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.1 | 0.3 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 1.6 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 1.1 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.1 | 0.7 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.3 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.1 | 0.7 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.2 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.1 | 0.2 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.1 | 0.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.2 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.3 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.5 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.7 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.1 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.0 | 0.3 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.1 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.0 | 0.3 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.6 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.6 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.3 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.0 | 0.2 | GO:0032202 | telomere assembly(GO:0032202) scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.2 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0045083 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) response to diterpene(GO:1904629) cellular response to diterpene(GO:1904630) response to glucoside(GO:1904631) cellular response to glucoside(GO:1904632) |
| 0.0 | 0.4 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.4 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.2 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.5 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.4 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.4 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.3 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.2 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.2 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.2 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 0.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.0 | GO:0046108 | uridine catabolic process(GO:0006218) uridine metabolic process(GO:0046108) |
| 0.0 | 0.2 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.0 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 0.1 | GO:0021539 | subthalamus development(GO:0021539) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.1 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.8 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.9 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.7 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.7 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 1.8 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.3 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 0.2 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 0.7 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.6 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.5 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.6 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.4 | 1.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.3 | 0.9 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.2 | 0.9 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.2 | 0.6 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.6 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.4 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 0.6 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 0.6 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.7 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.6 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.5 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.3 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.1 | 0.7 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 1.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.3 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 0.4 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.5 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.2 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.4 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.8 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) proteasome binding(GO:0070628) |
| 0.0 | 0.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.4 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.2 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.2 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0042497 | triacyl lipopeptide binding(GO:0042497) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.8 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.6 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 1.7 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |