Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Zbtb49
Z-value: 0.16
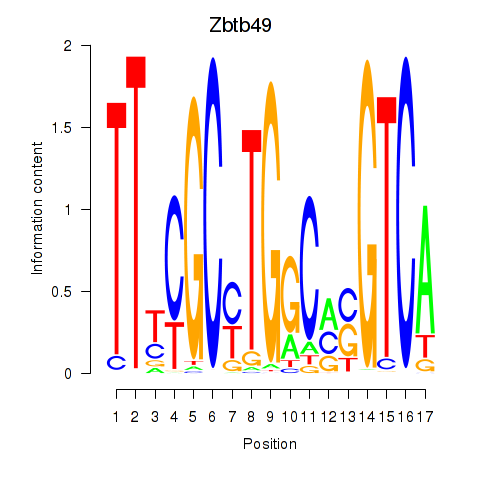
Transcription factors associated with Zbtb49
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb49
|
ENSMUSG00000029127.16 | zinc finger and BTB domain containing 49 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb49 | mm39_v1_chr5_-_38377746_38377801 | -0.08 | 9.0e-01 | Click! |
Activity profile of Zbtb49 motif
Sorted Z-values of Zbtb49 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_89099404 | 0.14 |
ENSMUST00000024916.7
|
Lhcgr
|
luteinizing hormone/choriogonadotropin receptor |
| chr1_+_32211792 | 0.12 |
ENSMUST00000027226.12
ENSMUST00000189878.2 ENSMUST00000188257.7 ENSMUST00000185666.2 |
Khdrbs2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr8_-_106723026 | 0.10 |
ENSMUST00000227363.2
ENSMUST00000081998.13 |
Dpep2
|
dipeptidase 2 |
| chr5_-_108777578 | 0.07 |
ENSMUST00000046603.15
|
Gak
|
cyclin G associated kinase |
| chr7_+_4463686 | 0.06 |
ENSMUST00000167298.2
ENSMUST00000171445.8 |
Eps8l1
|
EPS8-like 1 |
| chr15_-_102165884 | 0.06 |
ENSMUST00000043172.15
|
Rarg
|
retinoic acid receptor, gamma |
| chr5_+_108777636 | 0.04 |
ENSMUST00000146207.6
ENSMUST00000063272.13 |
Tmem175
|
transmembrane protein 175 |
| chr3_-_154302679 | 0.04 |
ENSMUST00000052774.8
ENSMUST00000170461.8 ENSMUST00000122976.2 |
Tyw3
|
tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) |
| chr17_+_24955613 | 0.03 |
ENSMUST00000115262.9
|
Msrb1
|
methionine sulfoxide reductase B1 |
| chr13_-_112717196 | 0.03 |
ENSMUST00000051756.8
|
Il31ra
|
interleukin 31 receptor A |
| chr15_-_102165740 | 0.03 |
ENSMUST00000135466.2
|
Rarg
|
retinoic acid receptor, gamma |
| chr8_-_106706035 | 0.02 |
ENSMUST00000034371.9
|
Dpep3
|
dipeptidase 3 |
| chrX_-_152479191 | 0.02 |
ENSMUST00000039424.14
ENSMUST00000112572.3 |
Kctd12b
|
potassium channel tetramerisation domain containing 12b |
| chr14_-_122703089 | 0.02 |
ENSMUST00000039118.7
|
Zic5
|
zinc finger protein of the cerebellum 5 |
| chr9_+_40180569 | 0.01 |
ENSMUST00000176185.8
|
Scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr9_+_40180726 | 0.01 |
ENSMUST00000171835.9
|
Scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr9_+_40180497 | 0.01 |
ENSMUST00000049941.12
|
Scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr7_-_97848688 | 0.01 |
ENSMUST00000098278.4
|
B3gnt6
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase) |
| chr2_-_80411724 | 0.01 |
ENSMUST00000111760.3
|
Nckap1
|
NCK-associated protein 1 |
| chr18_-_62044871 | 0.01 |
ENSMUST00000166783.3
ENSMUST00000049378.15 |
Ablim3
|
actin binding LIM protein family, member 3 |
| chr2_-_80411578 | 0.00 |
ENSMUST00000028386.12
|
Nckap1
|
NCK-associated protein 1 |
| chr2_-_167082505 | 0.00 |
ENSMUST00000088041.11
ENSMUST00000018113.8 |
Ptgis
|
prostaglandin I2 (prostacyclin) synthase |
| chr8_+_105727456 | 0.00 |
ENSMUST00000172032.4
|
Ces2h
|
carboxylesterase 2H |
| chr15_-_3333003 | 0.00 |
ENSMUST00000165386.2
|
Ccdc152
|
coiled-coil domain containing 152 |
| chr15_+_100321074 | 0.00 |
ENSMUST00000148928.2
|
Gm5475
|
predicted gene 5475 |
| chr7_-_139734637 | 0.00 |
ENSMUST00000059241.8
|
Sprn
|
shadow of prion protein |
| chr7_-_34914675 | 0.00 |
ENSMUST00000118444.3
ENSMUST00000122409.8 |
Lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr14_+_53279810 | 0.00 |
ENSMUST00000198439.5
ENSMUST00000103605.3 |
Trav8d-2
|
T cell receptor alpha variable 8D-2 |
| chr18_+_23886765 | 0.00 |
ENSMUST00000115830.8
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb49
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.0 | 0.1 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) Harderian gland development(GO:0070384) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |