Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
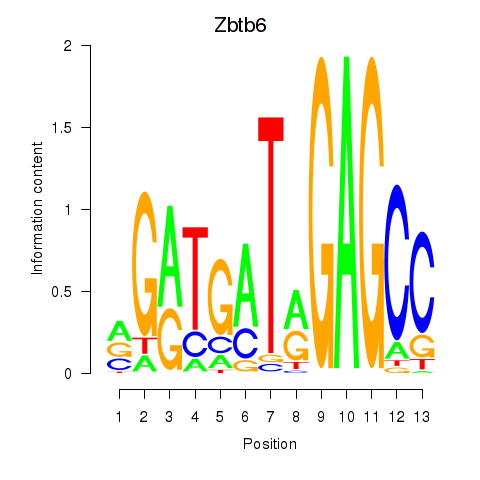
Results for Zbtb6
Z-value: 1.17
Transcription factors associated with Zbtb6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb6
|
ENSMUSG00000066798.4 | zinc finger and BTB domain containing 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb6 | mm39_v1_chr2_-_37333109_37333210 | 0.47 | 4.2e-01 | Click! |
Activity profile of Zbtb6 motif
Sorted Z-values of Zbtb6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_35658131 | 2.01 |
ENSMUST00000071951.14
ENSMUST00000116598.10 ENSMUST00000078205.14 ENSMUST00000076256.8 |
H2-Q7
|
histocompatibility 2, Q region locus 7 |
| chr17_+_35643818 | 1.74 |
ENSMUST00000174699.8
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr17_+_35643853 | 1.53 |
ENSMUST00000113879.4
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr4_-_87724533 | 0.83 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr4_-_87724512 | 0.65 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr11_+_96920751 | 0.58 |
ENSMUST00000021249.11
|
Scrn2
|
secernin 2 |
| chr1_+_127132712 | 0.57 |
ENSMUST00000038361.11
|
Mgat5
|
mannoside acetylglucosaminyltransferase 5 |
| chr4_+_129083553 | 0.54 |
ENSMUST00000106054.4
|
Yars
|
tyrosyl-tRNA synthetase |
| chr11_-_88742285 | 0.50 |
ENSMUST00000107903.8
|
Akap1
|
A kinase (PRKA) anchor protein 1 |
| chr15_-_79571977 | 0.48 |
ENSMUST00000023061.7
|
Josd1
|
Josephin domain containing 1 |
| chr17_+_35780977 | 0.48 |
ENSMUST00000174525.8
ENSMUST00000068291.7 |
H2-Q10
|
histocompatibility 2, Q region locus 10 |
| chr9_+_107468146 | 0.44 |
ENSMUST00000195746.2
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr15_+_102234802 | 0.43 |
ENSMUST00000169637.8
|
Pfdn5
|
prefoldin 5 |
| chr4_+_106418224 | 0.43 |
ENSMUST00000047973.4
|
Dhcr24
|
24-dehydrocholesterol reductase |
| chr8_-_95564881 | 0.42 |
ENSMUST00000034233.15
ENSMUST00000162538.9 |
Ciapin1
|
cytokine induced apoptosis inhibitor 1 |
| chr1_+_134110142 | 0.42 |
ENSMUST00000082060.10
ENSMUST00000153856.8 ENSMUST00000133701.8 ENSMUST00000132873.8 |
Chil1
|
chitinase-like 1 |
| chr15_+_78798116 | 0.41 |
ENSMUST00000089378.5
|
Pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase |
| chr15_+_103411689 | 0.40 |
ENSMUST00000226493.2
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent |
| chr8_-_61407760 | 0.39 |
ENSMUST00000110302.8
|
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr4_+_107659361 | 0.39 |
ENSMUST00000106731.4
|
Lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr3_+_96011810 | 0.38 |
ENSMUST00000132980.8
ENSMUST00000138206.8 ENSMUST00000090785.9 ENSMUST00000035519.12 |
Otud7b
|
OTU domain containing 7B |
| chrX_-_100310959 | 0.38 |
ENSMUST00000135038.2
|
Il2rg
|
interleukin 2 receptor, gamma chain |
| chr19_+_8748347 | 0.38 |
ENSMUST00000010248.4
|
Tmem223
|
transmembrane protein 223 |
| chr15_+_58004753 | 0.36 |
ENSMUST00000067563.9
|
Wdyhv1
|
WDYHV motif containing 1 |
| chr7_-_44785603 | 0.36 |
ENSMUST00000209467.2
|
Gm45713
|
predicted gene 45713 |
| chr12_-_69629758 | 0.35 |
ENSMUST00000058639.11
|
Vcpkmt
|
valosin containing protein lysine (K) methyltransferase |
| chr9_-_21671571 | 0.33 |
ENSMUST00000217382.2
ENSMUST00000214149.2 ENSMUST00000098942.6 ENSMUST00000216057.2 |
Spc24
|
SPC24, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr18_+_11972277 | 0.30 |
ENSMUST00000171109.9
ENSMUST00000046948.10 |
Cables1
|
CDK5 and Abl enzyme substrate 1 |
| chrX_+_149377416 | 0.30 |
ENSMUST00000112713.3
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr2_-_126460575 | 0.30 |
ENSMUST00000028838.5
|
Hdc
|
histidine decarboxylase |
| chr18_-_42395207 | 0.30 |
ENSMUST00000097590.5
|
Lars
|
leucyl-tRNA synthetase |
| chr19_+_45991907 | 0.29 |
ENSMUST00000099393.4
|
Hps6
|
HPS6, biogenesis of lysosomal organelles complex 2 subunit 3 |
| chr17_-_46464441 | 0.29 |
ENSMUST00000171172.3
|
Mad2l1bp
|
MAD2L1 binding protein |
| chr14_+_56640038 | 0.29 |
ENSMUST00000095793.3
ENSMUST00000223627.2 |
Rnf17
|
ring finger protein 17 |
| chr14_+_55132030 | 0.29 |
ENSMUST00000141446.8
ENSMUST00000139985.8 |
Pabpn1
|
poly(A) binding protein, nuclear 1 |
| chr7_-_37722938 | 0.29 |
ENSMUST00000206581.2
|
Uri1
|
URI1, prefoldin-like chaperone |
| chr4_+_63478478 | 0.28 |
ENSMUST00000080336.4
|
Tmem268
|
transmembrane protein 268 |
| chr5_-_107745443 | 0.28 |
ENSMUST00000100949.10
ENSMUST00000078021.13 |
Glmn
|
glomulin, FKBP associated protein |
| chr15_+_102381929 | 0.28 |
ENSMUST00000229746.2
ENSMUST00000230211.2 ENSMUST00000230539.2 |
Pcbp2
|
poly(rC) binding protein 2 |
| chr6_+_113600920 | 0.27 |
ENSMUST00000035673.8
|
Vhl
|
von Hippel-Lindau tumor suppressor |
| chr11_-_59397352 | 0.27 |
ENSMUST00000013262.15
ENSMUST00000101150.9 |
Zkscan17
|
zinc finger with KRAB and SCAN domains 17 |
| chr15_+_98006346 | 0.26 |
ENSMUST00000051226.8
|
Pfkm
|
phosphofructokinase, muscle |
| chr15_+_58004793 | 0.26 |
ENSMUST00000227142.2
ENSMUST00000226955.2 |
Wdyhv1
|
WDYHV motif containing 1 |
| chr5_-_33812830 | 0.25 |
ENSMUST00000200849.2
|
Tmem129
|
transmembrane protein 129 |
| chr11_+_74661647 | 0.24 |
ENSMUST00000010698.13
ENSMUST00000141755.8 |
Mettl16
|
methyltransferase like 16 |
| chr16_+_18695787 | 0.24 |
ENSMUST00000120532.9
ENSMUST00000004222.14 |
Hira
|
histone cell cycle regulator |
| chrX_-_156381638 | 0.23 |
ENSMUST00000112522.9
ENSMUST00000179062.8 |
Mbtps2
|
membrane-bound transcription factor peptidase, site 2 |
| chr10_+_79986280 | 0.22 |
ENSMUST00000153477.8
|
Midn
|
midnolin |
| chr4_-_42665763 | 0.22 |
ENSMUST00000238770.2
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr11_-_17903861 | 0.22 |
ENSMUST00000076661.7
|
Etaa1
|
Ewing tumor-associated antigen 1 |
| chr4_-_42168603 | 0.21 |
ENSMUST00000098121.4
|
Gm13305
|
predicted gene 13305 |
| chr11_-_54678433 | 0.21 |
ENSMUST00000153209.8
ENSMUST00000064104.13 |
Cdc42se2
|
CDC42 small effector 2 |
| chr8_+_72022300 | 0.21 |
ENSMUST00000212111.2
|
Slc27a1
|
solute carrier family 27 (fatty acid transporter), member 1 |
| chr5_+_135178509 | 0.21 |
ENSMUST00000153183.8
|
Tbl2
|
transducin (beta)-like 2 |
| chr4_-_107975723 | 0.21 |
ENSMUST00000030340.15
|
Scp2
|
sterol carrier protein 2, liver |
| chr3_+_87826834 | 0.21 |
ENSMUST00000137775.2
|
Mrpl24
|
mitochondrial ribosomal protein L24 |
| chr15_+_103411461 | 0.20 |
ENSMUST00000023132.5
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent |
| chr11_+_96920956 | 0.19 |
ENSMUST00000153482.2
|
Scrn2
|
secernin 2 |
| chr13_+_12580743 | 0.19 |
ENSMUST00000221560.2
ENSMUST00000071973.8 |
Ero1b
|
endoplasmic reticulum oxidoreductase 1 beta |
| chr15_+_102381705 | 0.19 |
ENSMUST00000229958.2
ENSMUST00000229184.2 ENSMUST00000230728.2 ENSMUST00000230114.2 |
Pcbp2
|
poly(rC) binding protein 2 |
| chr15_+_6329263 | 0.19 |
ENSMUST00000078019.13
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr1_-_135358228 | 0.19 |
ENSMUST00000041023.14
|
Ipo9
|
importin 9 |
| chr1_+_134109888 | 0.18 |
ENSMUST00000156873.8
|
Chil1
|
chitinase-like 1 |
| chr16_-_18052937 | 0.18 |
ENSMUST00000076957.7
|
Zdhhc8
|
zinc finger, DHHC domain containing 8 |
| chr19_-_8858247 | 0.17 |
ENSMUST00000096253.7
|
Uqcc3
|
ubiquinol-cytochrome c reductase complex assembly factor 3 |
| chr14_-_52258158 | 0.17 |
ENSMUST00000228580.2
ENSMUST00000226554.2 ENSMUST00000067549.15 |
Zfp219
|
zinc finger protein 219 |
| chr7_-_118455208 | 0.17 |
ENSMUST00000126792.9
ENSMUST00000116280.9 ENSMUST00000063607.12 |
Knop1
|
lysine rich nucleolar protein 1 |
| chr14_+_56905698 | 0.16 |
ENSMUST00000116468.2
|
Mphosph8
|
M-phase phosphoprotein 8 |
| chr12_+_111540920 | 0.16 |
ENSMUST00000075281.8
ENSMUST00000084953.13 |
Mark3
|
MAP/microtubule affinity regulating kinase 3 |
| chr1_+_74627506 | 0.16 |
ENSMUST00000113732.2
|
Bcs1l
|
BCS1-like (yeast) |
| chr8_-_13940234 | 0.16 |
ENSMUST00000033839.9
|
Coprs
|
coordinator of PRMT5, differentiation stimulator |
| chrX_-_84820209 | 0.16 |
ENSMUST00000142152.2
ENSMUST00000156390.8 |
Gk
|
glycerol kinase |
| chrX_-_153999333 | 0.16 |
ENSMUST00000112551.4
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr11_-_79971750 | 0.16 |
ENSMUST00000103233.10
ENSMUST00000061283.15 |
Crlf3
|
cytokine receptor-like factor 3 |
| chrX_+_49930311 | 0.16 |
ENSMUST00000114887.9
|
Stk26
|
serine/threonine kinase 26 |
| chr9_+_108660989 | 0.16 |
ENSMUST00000192307.6
ENSMUST00000193560.6 ENSMUST00000194875.6 |
Ip6k2
|
inositol hexaphosphate kinase 2 |
| chr17_-_3608056 | 0.15 |
ENSMUST00000041003.8
|
Tfb1m
|
transcription factor B1, mitochondrial |
| chr2_-_30084124 | 0.15 |
ENSMUST00000113659.8
ENSMUST00000113660.2 |
Kyat1
|
kynurenine aminotransferase 1 |
| chr2_+_78699360 | 0.15 |
ENSMUST00000028398.14
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr8_-_23295603 | 0.15 |
ENSMUST00000163739.3
ENSMUST00000210656.2 |
Ap3m2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr1_+_58841808 | 0.15 |
ENSMUST00000190213.2
|
Casp8
|
caspase 8 |
| chr3_-_95014090 | 0.15 |
ENSMUST00000005768.8
ENSMUST00000107232.9 ENSMUST00000107236.9 |
Pip5k1a
|
phosphatidylinositol-4-phosphate 5-kinase, type 1 alpha |
| chr11_+_86375441 | 0.15 |
ENSMUST00000020827.7
|
Rnft1
|
ring finger protein, transmembrane 1 |
| chr2_+_144441817 | 0.15 |
ENSMUST00000028917.7
|
Dtd1
|
D-tyrosyl-tRNA deacylase 1 |
| chr5_-_107745404 | 0.15 |
ENSMUST00000124140.2
|
Glmn
|
glomulin, FKBP associated protein |
| chr13_-_22193900 | 0.14 |
ENSMUST00000006341.4
|
Prss16
|
protease, serine 16 (thymus) |
| chr11_+_43572825 | 0.14 |
ENSMUST00000061070.6
ENSMUST00000094294.5 |
Pwwp2a
|
PWWP domain containing 2A |
| chr17_+_56316305 | 0.14 |
ENSMUST00000159340.8
|
Mpnd
|
MPN domain containing |
| chr7_-_118455477 | 0.13 |
ENSMUST00000106550.11
|
Knop1
|
lysine rich nucleolar protein 1 |
| chr7_+_126832399 | 0.13 |
ENSMUST00000056232.7
|
Zfp553
|
zinc finger protein 553 |
| chr10_+_81104006 | 0.13 |
ENSMUST00000057798.9
ENSMUST00000219304.2 |
Apba3
|
amyloid beta (A4) precursor protein-binding, family A, member 3 |
| chr19_+_3372296 | 0.12 |
ENSMUST00000237938.2
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr15_-_89361571 | 0.12 |
ENSMUST00000165199.8
|
Arsa
|
arylsulfatase A |
| chr3_+_86131970 | 0.12 |
ENSMUST00000192145.6
ENSMUST00000194759.6 ENSMUST00000107635.7 |
Lrba
|
LPS-responsive beige-like anchor |
| chr17_-_34250616 | 0.12 |
ENSMUST00000169397.9
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr17_+_29171386 | 0.12 |
ENSMUST00000118762.9
ENSMUST00000057174.16 ENSMUST00000232874.2 ENSMUST00000232772.2 ENSMUST00000233334.2 ENSMUST00000150858.2 ENSMUST00000233064.2 |
Kctd20
|
potassium channel tetramerisation domain containing 20 |
| chr10_-_40018243 | 0.12 |
ENSMUST00000092566.8
ENSMUST00000213488.2 |
Slc16a10
|
solute carrier family 16 (monocarboxylic acid transporters), member 10 |
| chr8_-_106434565 | 0.12 |
ENSMUST00000013299.11
|
Enkd1
|
enkurin domain containing 1 |
| chr14_+_55132061 | 0.12 |
ENSMUST00000172557.2
|
Pabpn1
|
poly(A) binding protein, nuclear 1 |
| chr3_-_95014218 | 0.12 |
ENSMUST00000107233.9
|
Pip5k1a
|
phosphatidylinositol-4-phosphate 5-kinase, type 1 alpha |
| chr19_+_4242064 | 0.11 |
ENSMUST00000046094.6
|
Ppp1ca
|
protein phosphatase 1 catalytic subunit alpha |
| chr1_+_121358778 | 0.11 |
ENSMUST00000036025.16
ENSMUST00000112621.2 |
Ccdc93
|
coiled-coil domain containing 93 |
| chr8_+_95564949 | 0.11 |
ENSMUST00000034234.15
ENSMUST00000159871.4 |
Coq9
|
coenzyme Q9 |
| chr11_+_80074677 | 0.11 |
ENSMUST00000017839.3
|
Rnf135
|
ring finger protein 135 |
| chr19_-_5507532 | 0.11 |
ENSMUST00000236881.2
|
Ccdc85b
|
coiled-coil domain containing 85B |
| chr18_-_42395131 | 0.11 |
ENSMUST00000236102.2
|
Lars
|
leucyl-tRNA synthetase |
| chr14_-_69944942 | 0.11 |
ENSMUST00000121142.4
|
R3hcc1
|
R3H domain and coiled-coil containing 1 |
| chrX_+_47001264 | 0.10 |
ENSMUST00000001202.15
ENSMUST00000115020.8 |
Ocrl
|
OCRL, inositol polyphosphate-5-phosphatase |
| chr17_+_56316201 | 0.10 |
ENSMUST00000149441.8
ENSMUST00000162883.8 ENSMUST00000159996.8 |
Mpnd
|
MPN domain containing |
| chr4_-_156285247 | 0.10 |
ENSMUST00000085425.6
|
Isg15
|
ISG15 ubiquitin-like modifier |
| chr8_+_94899292 | 0.10 |
ENSMUST00000034214.8
ENSMUST00000212806.2 |
Mt2
|
metallothionein 2 |
| chr3_-_95214443 | 0.10 |
ENSMUST00000015846.9
|
Anxa9
|
annexin A9 |
| chr5_+_107745626 | 0.10 |
ENSMUST00000112655.8
|
Rpap2
|
RNA polymerase II associated protein 2 |
| chr4_-_129083335 | 0.10 |
ENSMUST00000106061.9
ENSMUST00000072431.13 |
S100pbp
|
S100P binding protein |
| chrX_-_84820250 | 0.10 |
ENSMUST00000113978.9
|
Gk
|
glycerol kinase |
| chr13_-_85437201 | 0.10 |
ENSMUST00000223598.2
|
Rasa1
|
RAS p21 protein activator 1 |
| chr6_+_113508636 | 0.10 |
ENSMUST00000036340.12
ENSMUST00000204827.3 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr13_-_85437118 | 0.09 |
ENSMUST00000109552.3
|
Rasa1
|
RAS p21 protein activator 1 |
| chr17_+_35069347 | 0.09 |
ENSMUST00000097343.11
ENSMUST00000173357.8 ENSMUST00000173065.8 ENSMUST00000165953.3 |
Nelfe
|
negative elongation factor complex member E, Rdbp |
| chr13_+_110531571 | 0.09 |
ENSMUST00000022212.9
|
Plk2
|
polo like kinase 2 |
| chr17_-_36206812 | 0.09 |
ENSMUST00000059740.15
|
2310061I04Rik
|
RIKEN cDNA 2310061I04 gene |
| chr4_+_63478454 | 0.09 |
ENSMUST00000124332.8
ENSMUST00000150360.8 |
Tmem268
|
transmembrane protein 268 |
| chr6_-_113696809 | 0.09 |
ENSMUST00000203770.3
ENSMUST00000064993.8 |
Ghrl
|
ghrelin |
| chr5_+_107745562 | 0.09 |
ENSMUST00000112651.8
ENSMUST00000112654.8 ENSMUST00000065422.12 |
Rpap2
|
RNA polymerase II associated protein 2 |
| chr8_-_106074585 | 0.09 |
ENSMUST00000014922.5
|
Fhod1
|
formin homology 2 domain containing 1 |
| chr5_-_72325482 | 0.09 |
ENSMUST00000196241.2
ENSMUST00000013693.11 |
Commd8
|
COMM domain containing 8 |
| chr7_+_126832215 | 0.09 |
ENSMUST00000106312.4
|
Zfp553
|
zinc finger protein 553 |
| chr17_+_29171655 | 0.08 |
ENSMUST00000117672.9
ENSMUST00000153831.9 |
Kctd20
|
potassium channel tetramerisation domain containing 20 |
| chr7_-_118454836 | 0.08 |
ENSMUST00000106549.9
ENSMUST00000152309.3 ENSMUST00000033277.9 |
Knop1
|
lysine rich nucleolar protein 1 |
| chr4_+_117692583 | 0.08 |
ENSMUST00000169885.8
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr19_-_10282218 | 0.08 |
ENSMUST00000039327.11
|
Dagla
|
diacylglycerol lipase, alpha |
| chr4_-_140780972 | 0.07 |
ENSMUST00000040222.14
|
Crocc
|
ciliary rootlet coiled-coil, rootletin |
| chr15_+_6329278 | 0.07 |
ENSMUST00000159046.2
ENSMUST00000161040.8 |
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr14_-_57635969 | 0.07 |
ENSMUST00000022517.9
|
Cryl1
|
crystallin, lambda 1 |
| chr10_-_82458760 | 0.07 |
ENSMUST00000176200.8
ENSMUST00000183416.2 |
1190007I07Rik
|
RIKEN cDNA 1190007I07 gene |
| chr19_+_38825120 | 0.07 |
ENSMUST00000235558.2
|
Tbc1d12
|
TBC1D12: TBC1 domain family, member 12 |
| chr1_-_74627264 | 0.07 |
ENSMUST00000066986.13
|
Zfp142
|
zinc finger protein 142 |
| chr16_-_26190578 | 0.07 |
ENSMUST00000023154.3
|
Cldn1
|
claudin 1 |
| chr11_-_100510423 | 0.07 |
ENSMUST00000137688.9
ENSMUST00000239389.2 ENSMUST00000155152.9 ENSMUST00000154972.9 ENSMUST00000239479.2 ENSMUST00000132886.3 |
Dnajc7
|
DnaJ heat shock protein family (Hsp40) member C7 |
| chr14_+_55131568 | 0.07 |
ENSMUST00000116476.9
ENSMUST00000022808.14 ENSMUST00000150975.8 |
Pabpn1
|
poly(A) binding protein, nuclear 1 |
| chr9_-_21982637 | 0.07 |
ENSMUST00000179605.9
ENSMUST00000043922.7 |
Zfp653
|
zinc finger protein 653 |
| chr13_+_3588063 | 0.06 |
ENSMUST00000223396.2
ENSMUST00000059515.8 ENSMUST00000222365.2 |
Gdi2
|
guanosine diphosphate (GDP) dissociation inhibitor 2 |
| chr16_-_18165876 | 0.06 |
ENSMUST00000125287.9
|
Tango2
|
transport and golgi organization 2 |
| chr7_+_24246575 | 0.06 |
ENSMUST00000063249.9
|
Xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr3_-_129548954 | 0.06 |
ENSMUST00000029653.7
|
Egf
|
epidermal growth factor |
| chr15_+_58004831 | 0.06 |
ENSMUST00000226889.2
|
Wdyhv1
|
WDYHV motif containing 1 |
| chr10_-_120815232 | 0.06 |
ENSMUST00000119944.8
ENSMUST00000119093.2 |
Lemd3
|
LEM domain containing 3 |
| chr17_+_35561218 | 0.06 |
ENSMUST00000074806.12
|
H2-Q2
|
histocompatibility 2, Q region locus 2 |
| chr7_+_105024231 | 0.06 |
ENSMUST00000106805.4
|
Gm5901
|
predicted gene 5901 |
| chr7_+_28488380 | 0.06 |
ENSMUST00000209035.2
ENSMUST00000059857.8 |
Rinl
|
Ras and Rab interactor-like |
| chr14_+_79718604 | 0.05 |
ENSMUST00000040131.13
|
Elf1
|
E74-like factor 1 |
| chr11_-_100510992 | 0.05 |
ENSMUST00000014339.15
ENSMUST00000239490.2 ENSMUST00000239410.2 |
Dnajc7
|
DnaJ heat shock protein family (Hsp40) member C7 |
| chr4_+_103000248 | 0.05 |
ENSMUST00000106855.2
|
Mier1
|
MEIR1 treanscription regulator |
| chr15_+_99023775 | 0.05 |
ENSMUST00000230628.2
ENSMUST00000229073.2 ENSMUST00000229147.2 |
Spats2
|
spermatogenesis associated, serine-rich 2 |
| chr4_-_140781015 | 0.05 |
ENSMUST00000102491.10
|
Crocc
|
ciliary rootlet coiled-coil, rootletin |
| chr10_-_95509251 | 0.05 |
ENSMUST00000218771.2
ENSMUST00000099328.2 |
Anapc15-ps
|
anaphase promoting complex C subunit 15, pseudogene |
| chr18_-_53877579 | 0.05 |
ENSMUST00000049811.8
ENSMUST00000237062.2 |
Cep120
|
centrosomal protein 120 |
| chr13_+_55517545 | 0.05 |
ENSMUST00000063771.14
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr14_-_69945022 | 0.05 |
ENSMUST00000118374.8
|
R3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr11_+_3939924 | 0.05 |
ENSMUST00000109981.2
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr2_+_90948481 | 0.05 |
ENSMUST00000137942.8
ENSMUST00000111430.10 ENSMUST00000169776.2 |
Mybpc3
|
myosin binding protein C, cardiac |
| chr6_-_113508536 | 0.04 |
ENSMUST00000032425.7
|
Emc3
|
ER membrane protein complex subunit 3 |
| chr17_+_37180437 | 0.04 |
ENSMUST00000060524.11
|
Trim10
|
tripartite motif-containing 10 |
| chr10_+_81104041 | 0.04 |
ENSMUST00000218742.2
|
Apba3
|
amyloid beta (A4) precursor protein-binding, family A, member 3 |
| chr9_+_113760376 | 0.04 |
ENSMUST00000214095.2
ENSMUST00000116492.3 ENSMUST00000216558.2 |
Ubp1
|
upstream binding protein 1 |
| chr10_-_75768302 | 0.04 |
ENSMUST00000120281.8
ENSMUST00000000924.13 |
Mmp11
|
matrix metallopeptidase 11 |
| chr1_-_74627123 | 0.04 |
ENSMUST00000027315.14
ENSMUST00000113737.8 |
Zfp142
|
zinc finger protein 142 |
| chr6_-_60806810 | 0.04 |
ENSMUST00000163779.8
|
Snca
|
synuclein, alpha |
| chr3_-_100396635 | 0.04 |
ENSMUST00000061455.9
|
Tent5c
|
terminal nucleotidyltransferase 5C |
| chr3_-_145355725 | 0.04 |
ENSMUST00000029846.5
|
Ccn1
|
cellular communication network factor 1 |
| chr15_+_99024394 | 0.04 |
ENSMUST00000063517.6
|
Spats2
|
spermatogenesis associated, serine-rich 2 |
| chr11_+_74661734 | 0.03 |
ENSMUST00000128504.2
|
Mettl16
|
methyltransferase like 16 |
| chr15_-_93173032 | 0.03 |
ENSMUST00000057896.5
ENSMUST00000049484.13 ENSMUST00000230063.2 |
Gxylt1
|
glucoside xylosyltransferase 1 |
| chr7_-_97827461 | 0.03 |
ENSMUST00000040971.14
|
Capn5
|
calpain 5 |
| chr3_+_108000425 | 0.03 |
ENSMUST00000151326.8
|
Gnat2
|
guanine nucleotide binding protein, alpha transducing 2 |
| chr6_-_113696390 | 0.03 |
ENSMUST00000203588.2
ENSMUST00000204163.3 ENSMUST00000203363.3 |
Ghrl
|
ghrelin |
| chr11_-_98084086 | 0.03 |
ENSMUST00000092735.12
ENSMUST00000107545.9 |
Med1
|
mediator complex subunit 1 |
| chr2_-_92201342 | 0.03 |
ENSMUST00000176810.8
ENSMUST00000090582.11 ENSMUST00000068586.13 |
Large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr17_+_88837540 | 0.03 |
ENSMUST00000038551.8
|
Ppp1r21
|
protein phosphatase 1, regulatory subunit 21 |
| chr16_+_18066825 | 0.03 |
ENSMUST00000100099.10
|
Trmt2a
|
TRM2 tRNA methyltransferase 2A |
| chr16_+_18066730 | 0.02 |
ENSMUST00000115640.8
ENSMUST00000140206.8 |
Trmt2a
|
TRM2 tRNA methyltransferase 2A |
| chr7_+_118454957 | 0.02 |
ENSMUST00000208658.2
ENSMUST00000098087.9 ENSMUST00000106547.2 |
Iqck
|
IQ motif containing K |
| chr11_+_9068507 | 0.02 |
ENSMUST00000164791.8
ENSMUST00000130522.8 |
Upp1
|
uridine phosphorylase 1 |
| chr10_-_82459024 | 0.02 |
ENSMUST00000183363.2
ENSMUST00000079648.12 ENSMUST00000185168.8 |
1190007I07Rik
|
RIKEN cDNA 1190007I07 gene |
| chr16_-_19079594 | 0.02 |
ENSMUST00000103752.3
ENSMUST00000197518.2 |
Iglv2
|
immunoglobulin lambda variable 2 |
| chr11_-_51153870 | 0.02 |
ENSMUST00000054226.3
|
BC049762
|
cDNA sequence BC049762 |
| chr6_-_67931692 | 0.02 |
ENSMUST00000103313.3
|
Igkv9-123
|
immunoglobulin kappa variable 9-123 |
| chr11_-_102787950 | 0.02 |
ENSMUST00000067444.10
|
Gfap
|
glial fibrillary acidic protein |
| chr17_+_36134450 | 0.01 |
ENSMUST00000172846.2
|
Flot1
|
flotillin 1 |
| chr18_+_37488174 | 0.01 |
ENSMUST00000192867.2
ENSMUST00000051163.3 |
Pcdhb8
|
protocadherin beta 8 |
| chr8_-_86091946 | 0.01 |
ENSMUST00000034133.14
|
Mylk3
|
myosin light chain kinase 3 |
| chr14_+_14475188 | 0.01 |
ENSMUST00000026315.8
|
Dnase1l3
|
deoxyribonuclease 1-like 3 |
| chr16_+_17095416 | 0.01 |
ENSMUST00000232364.2
|
Tmem191c
|
transmembrane protein 191C |
| chr11_-_102787972 | 0.01 |
ENSMUST00000077902.5
|
Gfap
|
glial fibrillary acidic protein |
| chr4_-_129083439 | 0.01 |
ENSMUST00000106059.8
|
S100pbp
|
S100P binding protein |
| chr11_-_73377648 | 0.01 |
ENSMUST00000078358.2
|
Olfr381
|
olfactory receptor 381 |
| chr12_-_116226565 | 0.01 |
ENSMUST00000039349.8
|
Dync2i1
|
dynein 2 intermediate chain 1 |
| chrX_-_52328963 | 0.01 |
ENSMUST00000074861.9
|
Plac1
|
placental specific protein 1 |
| chr7_+_3439144 | 0.01 |
ENSMUST00000182222.8
|
Cacng8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr18_+_37646674 | 0.01 |
ENSMUST00000061405.6
|
Pcdhb21
|
protocadherin beta 21 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.1 | 0.4 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.1 | 0.4 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 0.5 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 5.6 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 0.3 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.1 | 1.7 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.2 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.1 | 0.3 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.1 | 0.4 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.4 | GO:0032831 | positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.1 | 0.3 | GO:0006548 | histamine metabolic process(GO:0001692) histidine catabolic process(GO:0006548) |
| 0.1 | 0.5 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.2 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 0.3 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.1 | 0.2 | GO:0032379 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.1 | 0.2 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.0 | 0.4 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.6 | GO:0036005 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.0 | 0.2 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.2 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:1904000 | positive regulation of eating behavior(GO:1904000) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) negative regulation of energy homeostasis(GO:2000506) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.3 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.1 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.4 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.5 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.4 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.4 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.3 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.1 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.0 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.0 | 0.2 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.2 | GO:0036119 | response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.3 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 1.0 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.7 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.0 | 0.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 1.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.5 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.8 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.2 | 0.7 | GO:0070773 | protein-N-terminal glutamine amidohydrolase activity(GO:0070773) |
| 0.1 | 0.6 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.4 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 0.4 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 0.1 | 0.4 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.1 | 0.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.4 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.1 | 0.3 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.1 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.4 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.1 | 0.3 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.1 | 0.6 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.6 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.2 | GO:1904121 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.1 | 0.2 | GO:0047316 | L-phenylalanine:pyruvate aminotransferase activity(GO:0047312) glutamine-phenylpyruvate transaminase activity(GO:0047316) L-glutamine:pyruvate aminotransferase activity(GO:0047945) |
| 0.0 | 0.6 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.1 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.2 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.8 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.5 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.5 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.4 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.2 | GO:0052723 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.0 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.0 | 0.1 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.6 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |