Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Zbtb7a
Z-value: 0.55
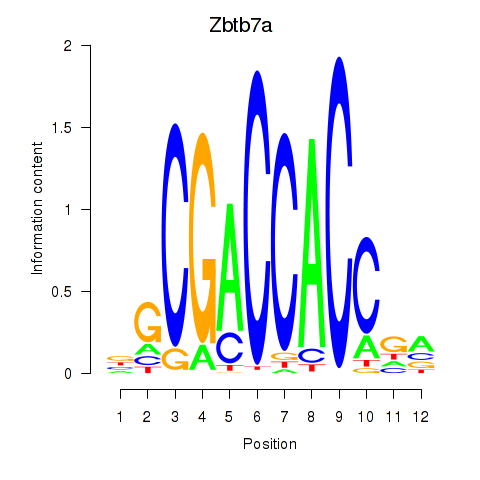
Transcription factors associated with Zbtb7a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb7a
|
ENSMUSG00000035011.16 | zinc finger and BTB domain containing 7a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb7a | mm39_v1_chr10_+_80972089_80972105 | 0.74 | 1.6e-01 | Click! |
Activity profile of Zbtb7a motif
Sorted Z-values of Zbtb7a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_71318353 | 0.78 |
ENSMUST00000064780.4
|
Gabre
|
gamma-aminobutyric acid (GABA) A receptor, subunit epsilon |
| chr7_-_15781838 | 0.42 |
ENSMUST00000210781.2
|
Bicra
|
BRD4 interacting chromatin remodeling complex associated protein |
| chr6_+_6863269 | 0.33 |
ENSMUST00000171311.8
ENSMUST00000160937.9 |
Dlx6
|
distal-less homeobox 6 |
| chr15_+_78784043 | 0.32 |
ENSMUST00000001226.11
|
Sh3bp1
|
SH3-domain binding protein 1 |
| chr9_+_100525807 | 0.31 |
ENSMUST00000133388.2
|
Stag1
|
stromal antigen 1 |
| chr13_-_32967937 | 0.30 |
ENSMUST00000238977.3
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr2_-_143853122 | 0.30 |
ENSMUST00000016072.12
ENSMUST00000037875.6 |
Rrbp1
|
ribosome binding protein 1 |
| chr11_+_68322945 | 0.30 |
ENSMUST00000021283.8
|
Pik3r5
|
phosphoinositide-3-kinase regulatory subunit 5 |
| chr4_-_123558516 | 0.29 |
ENSMUST00000147030.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr15_-_76702170 | 0.27 |
ENSMUST00000175843.3
ENSMUST00000177026.3 ENSMUST00000176736.3 ENSMUST00000036176.16 ENSMUST00000176219.9 ENSMUST00000239134.2 ENSMUST00000239003.2 ENSMUST00000077821.10 |
Arhgap39
|
Rho GTPase activating protein 39 |
| chr15_-_98465516 | 0.27 |
ENSMUST00000012104.7
|
Ccnt1
|
cyclin T1 |
| chr11_+_115078299 | 0.27 |
ENSMUST00000100235.9
ENSMUST00000061450.7 |
Tmem104
|
transmembrane protein 104 |
| chr8_+_91635192 | 0.25 |
ENSMUST00000211403.2
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr10_-_12744025 | 0.24 |
ENSMUST00000219660.2
|
Utrn
|
utrophin |
| chr12_-_76756772 | 0.23 |
ENSMUST00000166101.2
|
Sptb
|
spectrin beta, erythrocytic |
| chr3_+_63203235 | 0.23 |
ENSMUST00000194134.6
|
Mme
|
membrane metallo endopeptidase |
| chr1_+_156386327 | 0.22 |
ENSMUST00000173929.8
|
Abl2
|
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
| chr2_-_25873068 | 0.22 |
ENSMUST00000127823.2
ENSMUST00000134882.8 |
Camsap1
|
calmodulin regulated spectrin-associated protein 1 |
| chr11_-_70578775 | 0.20 |
ENSMUST00000036299.14
ENSMUST00000119120.2 ENSMUST00000100933.10 |
Camta2
|
calmodulin binding transcription activator 2 |
| chrX_+_7439839 | 0.20 |
ENSMUST00000144719.9
ENSMUST00000234896.2 |
Flicr
Foxp3
|
Foxp3 regulating long intergenic noncoding RNA forkhead box P3 |
| chr4_+_137004793 | 0.20 |
ENSMUST00000045747.5
|
Wnt4
|
wingless-type MMTV integration site family, member 4 |
| chr19_+_10502679 | 0.19 |
ENSMUST00000235674.2
|
Cpsf7
|
cleavage and polyadenylation specific factor 7 |
| chr10_-_49664839 | 0.19 |
ENSMUST00000220263.2
ENSMUST00000218823.2 |
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2) |
| chr11_+_70453724 | 0.18 |
ENSMUST00000102559.11
|
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr10_-_12743915 | 0.18 |
ENSMUST00000219584.2
|
Utrn
|
utrophin |
| chr19_+_10502612 | 0.18 |
ENSMUST00000237321.2
ENSMUST00000038379.5 |
Cpsf7
|
cleavage and polyadenylation specific factor 7 |
| chr11_+_70453666 | 0.18 |
ENSMUST00000072237.13
ENSMUST00000072873.14 |
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr5_+_108817133 | 0.18 |
ENSMUST00000119212.8
|
Idua
|
iduronidase, alpha-L |
| chr4_+_132903646 | 0.18 |
ENSMUST00000105912.2
|
Wasf2
|
WASP family, member 2 |
| chr11_-_70578744 | 0.17 |
ENSMUST00000108545.9
ENSMUST00000120261.8 |
Camta2
|
calmodulin binding transcription activator 2 |
| chr11_+_117545037 | 0.17 |
ENSMUST00000026658.13
|
Tnrc6c
|
trinucleotide repeat containing 6C |
| chr2_-_32271833 | 0.17 |
ENSMUST00000146423.2
|
1110008P14Rik
|
RIKEN cDNA 1110008P14 gene |
| chr15_+_78783867 | 0.16 |
ENSMUST00000134703.8
ENSMUST00000061239.14 ENSMUST00000109698.9 |
Gm49510
Sh3bp1
|
predicted gene, 49510 SH3-domain binding protein 1 |
| chr5_-_144294854 | 0.16 |
ENSMUST00000055190.8
|
Baiap2l1
|
BAI1-associated protein 2-like 1 |
| chr1_+_131936022 | 0.16 |
ENSMUST00000146432.2
|
Elk4
|
ELK4, member of ETS oncogene family |
| chrX_+_55391749 | 0.16 |
ENSMUST00000101560.4
|
Zfp449
|
zinc finger protein 449 |
| chr1_-_170417354 | 0.15 |
ENSMUST00000160456.8
|
Nos1ap
|
nitric oxide synthase 1 (neuronal) adaptor protein |
| chr19_-_61215743 | 0.15 |
ENSMUST00000237386.2
|
Csf2ra
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr12_+_116041340 | 0.15 |
ENSMUST00000011315.10
|
Vipr2
|
vasoactive intestinal peptide receptor 2 |
| chr7_-_6699422 | 0.15 |
ENSMUST00000122432.4
ENSMUST00000002336.16 |
Zim1
|
zinc finger, imprinted 1 |
| chr13_-_38221045 | 0.15 |
ENSMUST00000089840.5
|
Cage1
|
cancer antigen 1 |
| chr11_-_97877219 | 0.15 |
ENSMUST00000107565.3
ENSMUST00000107564.2 ENSMUST00000017561.15 |
Plxdc1
|
plexin domain containing 1 |
| chr2_-_69416365 | 0.15 |
ENSMUST00000100051.9
ENSMUST00000092551.5 ENSMUST00000080953.12 |
Lrp2
|
low density lipoprotein receptor-related protein 2 |
| chr11_+_117545618 | 0.15 |
ENSMUST00000106344.8
|
Tnrc6c
|
trinucleotide repeat containing 6C |
| chr14_-_19261196 | 0.15 |
ENSMUST00000112797.12
ENSMUST00000225885.2 |
D830030K20Rik
|
RIKEN cDNA D830030K20 gene |
| chrX_+_55777139 | 0.14 |
ENSMUST00000023854.10
ENSMUST00000114769.9 |
Fhl1
|
four and a half LIM domains 1 |
| chr9_-_89505178 | 0.14 |
ENSMUST00000044491.13
|
Minar1
|
membrane integral NOTCH2 associated receptor 1 |
| chr16_-_4376471 | 0.14 |
ENSMUST00000230875.2
|
Tfap4
|
transcription factor AP4 |
| chr8_-_106198112 | 0.14 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr9_+_55234197 | 0.14 |
ENSMUST00000085754.10
ENSMUST00000034862.5 |
Tmem266
|
transmembrane protein 266 |
| chr12_+_108145997 | 0.14 |
ENSMUST00000101055.5
|
Ccnk
|
cyclin K |
| chr2_-_160714904 | 0.14 |
ENSMUST00000109460.8
ENSMUST00000127201.2 |
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr14_+_6226418 | 0.14 |
ENSMUST00000112625.9
|
Oxsm
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr19_-_10502468 | 0.14 |
ENSMUST00000025570.8
ENSMUST00000236455.2 |
Sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr12_-_108668520 | 0.14 |
ENSMUST00000167978.9
ENSMUST00000021691.6 |
Degs2
|
delta(4)-desaturase, sphingolipid 2 |
| chr15_+_76131020 | 0.13 |
ENSMUST00000229380.2
|
Grina
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr7_+_27879650 | 0.13 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr2_-_25873189 | 0.13 |
ENSMUST00000114167.9
ENSMUST00000091268.11 ENSMUST00000183461.8 |
Camsap1
|
calmodulin regulated spectrin-associated protein 1 |
| chr8_-_64659004 | 0.13 |
ENSMUST00000066166.6
|
Tll1
|
tolloid-like |
| chr11_+_70453806 | 0.13 |
ENSMUST00000079244.12
ENSMUST00000102558.11 |
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr11_+_68582923 | 0.13 |
ENSMUST00000018887.15
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr6_+_83055321 | 0.13 |
ENSMUST00000165164.9
ENSMUST00000092614.9 |
Pcgf1
|
polycomb group ring finger 1 |
| chr9_+_86454018 | 0.13 |
ENSMUST00000185566.7
ENSMUST00000034988.10 ENSMUST00000179212.3 |
Rwdd2a
|
RWD domain containing 2A |
| chr1_+_55127110 | 0.13 |
ENSMUST00000075242.7
|
Hspe1
|
heat shock protein 1 (chaperonin 10) |
| chr16_+_75389732 | 0.13 |
ENSMUST00000046378.14
ENSMUST00000114249.8 ENSMUST00000114253.2 |
Rbm11
|
RNA binding motif protein 11 |
| chr12_+_108145802 | 0.13 |
ENSMUST00000221167.2
|
Ccnk
|
cyclin K |
| chr19_-_5135510 | 0.13 |
ENSMUST00000140389.8
ENSMUST00000151413.2 ENSMUST00000077066.8 |
Tmem151a
|
transmembrane protein 151A |
| chr18_-_46345661 | 0.13 |
ENSMUST00000037011.6
|
Trim36
|
tripartite motif-containing 36 |
| chr11_+_68582731 | 0.12 |
ENSMUST00000102611.10
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr3_-_129854477 | 0.12 |
ENSMUST00000001079.15
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae) |
| chr4_-_128699838 | 0.12 |
ENSMUST00000106072.9
ENSMUST00000170934.3 |
Zfp362
|
zinc finger protein 362 |
| chr3_-_146205429 | 0.12 |
ENSMUST00000029839.11
|
Spata1
|
spermatogenesis associated 1 |
| chr9_+_124196337 | 0.12 |
ENSMUST00000188509.2
|
Ppp2r3d
|
protein phosphatase 2 (formerly 2A), regulatory subunit B'', delta |
| chr19_-_5323092 | 0.12 |
ENSMUST00000237463.2
ENSMUST00000025786.9 |
Pacs1
|
phosphofurin acidic cluster sorting protein 1 |
| chr2_-_90410922 | 0.12 |
ENSMUST00000168621.3
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr17_+_34341766 | 0.12 |
ENSMUST00000042121.11
|
H2-DMa
|
histocompatibility 2, class II, locus DMa |
| chr11_+_120612278 | 0.12 |
ENSMUST00000018156.12
|
Rac3
|
Rac family small GTPase 3 |
| chr3_-_32670628 | 0.12 |
ENSMUST00000193050.2
ENSMUST00000108234.8 ENSMUST00000155737.8 |
Gnb4
|
guanine nucleotide binding protein (G protein), beta 4 |
| chr4_+_152171286 | 0.12 |
ENSMUST00000118648.8
|
Plekhg5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr7_+_12615091 | 0.11 |
ENSMUST00000144578.2
|
Zfp128
|
zinc finger protein 128 |
| chr2_+_167774247 | 0.11 |
ENSMUST00000029053.8
|
Ptpn1
|
protein tyrosine phosphatase, non-receptor type 1 |
| chr11_-_74480870 | 0.11 |
ENSMUST00000145524.2
ENSMUST00000102521.9 |
Rap1gap2
|
RAP1 GTPase activating protein 2 |
| chr2_-_37312881 | 0.11 |
ENSMUST00000112936.4
ENSMUST00000112934.8 |
Rc3h2
|
ring finger and CCCH-type zinc finger domains 2 |
| chr11_-_70578905 | 0.11 |
ENSMUST00000108544.8
|
Camta2
|
calmodulin binding transcription activator 2 |
| chr10_+_79763164 | 0.11 |
ENSMUST00000105376.2
|
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr7_+_135139542 | 0.11 |
ENSMUST00000073961.8
|
Ptpre
|
protein tyrosine phosphatase, receptor type, E |
| chr19_+_23736205 | 0.11 |
ENSMUST00000025830.9
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr3_+_146205864 | 0.11 |
ENSMUST00000119130.2
|
Gng5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr7_-_28135616 | 0.11 |
ENSMUST00000208199.2
|
Samd4b
|
sterile alpha motif domain containing 4B |
| chr2_+_128971620 | 0.11 |
ENSMUST00000035481.5
|
Chchd5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr17_-_66384017 | 0.11 |
ENSMUST00000150766.2
ENSMUST00000038116.13 |
Ankrd12
|
ankyrin repeat domain 12 |
| chr17_+_46991972 | 0.11 |
ENSMUST00000002845.8
|
Mea1
|
male enhanced antigen 1 |
| chr18_+_34354031 | 0.11 |
ENSMUST00000115781.10
ENSMUST00000079362.13 |
Apc
|
APC, WNT signaling pathway regulator |
| chr8_+_61677253 | 0.10 |
ENSMUST00000209611.2
|
Sh3rf1
|
SH3 domain containing ring finger 1 |
| chr6_-_137626207 | 0.10 |
ENSMUST00000134630.6
ENSMUST00000058210.13 ENSMUST00000111878.8 |
Eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr11_+_115802828 | 0.10 |
ENSMUST00000132961.2
|
Smim6
|
small integral membrane protein 6 |
| chr2_-_160714473 | 0.10 |
ENSMUST00000103111.9
|
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr5_+_31107390 | 0.10 |
ENSMUST00000006814.9
|
Abhd1
|
abhydrolase domain containing 1 |
| chr2_-_156977032 | 0.10 |
ENSMUST00000139263.8
ENSMUST00000109549.3 ENSMUST00000088523.11 ENSMUST00000057725.10 |
Samhd1
|
SAM domain and HD domain, 1 |
| chr18_+_36098090 | 0.10 |
ENSMUST00000176873.8
ENSMUST00000177432.8 ENSMUST00000175734.2 |
Psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr11_-_107685383 | 0.10 |
ENSMUST00000021066.4
|
Cacng4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr7_-_73187369 | 0.10 |
ENSMUST00000172704.6
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr19_-_29782907 | 0.10 |
ENSMUST00000175726.8
|
9930021J03Rik
|
RIKEN cDNA 9930021J03 gene |
| chr1_-_155293075 | 0.10 |
ENSMUST00000027741.12
|
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr11_+_53410552 | 0.10 |
ENSMUST00000108987.8
ENSMUST00000121334.8 ENSMUST00000117061.8 |
Septin8
|
septin 8 |
| chr17_-_34341544 | 0.10 |
ENSMUST00000025193.14
|
Brd2
|
bromodomain containing 2 |
| chr6_-_116170389 | 0.10 |
ENSMUST00000088896.10
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr17_-_57366795 | 0.10 |
ENSMUST00000040280.14
|
Slc25a23
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23 |
| chr10_-_80578057 | 0.10 |
ENSMUST00000020420.9
|
Ap3d1
|
adaptor-related protein complex 3, delta 1 subunit |
| chr9_+_66257747 | 0.10 |
ENSMUST00000042824.13
|
Herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr15_+_78993111 | 0.09 |
ENSMUST00000040320.10
|
Micall1
|
microtubule associated monooxygenase, calponin and LIM domain containing -like 1 |
| chr1_-_155293141 | 0.09 |
ENSMUST00000111775.8
ENSMUST00000111774.2 |
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr2_-_165210622 | 0.09 |
ENSMUST00000141140.2
ENSMUST00000103085.8 |
Zfp663
|
zinc finger protein 663 |
| chr14_-_76348179 | 0.09 |
ENSMUST00000022585.5
|
Gpalpp1
|
GPALPP motifs containing 1 |
| chr9_+_102988940 | 0.09 |
ENSMUST00000189134.2
ENSMUST00000035155.8 |
Rab6b
|
RAB6B, member RAS oncogene family |
| chr9_+_67747668 | 0.09 |
ENSMUST00000077879.7
|
Vps13c
|
vacuolar protein sorting 13C |
| chr2_+_61423469 | 0.09 |
ENSMUST00000112494.2
|
Tank
|
TRAF family member-associated Nf-kappa B activator |
| chr7_+_28466160 | 0.09 |
ENSMUST00000122915.8
ENSMUST00000072965.5 ENSMUST00000170068.9 |
Sirt2
|
sirtuin 2 |
| chr8_-_26484131 | 0.09 |
ENSMUST00000061850.5
|
Pomk
|
protein-O-mannose kinase |
| chr17_+_35200823 | 0.09 |
ENSMUST00000114011.11
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr17_+_25798059 | 0.09 |
ENSMUST00000141606.3
ENSMUST00000063344.15 ENSMUST00000116641.9 |
Lmf1
|
lipase maturation factor 1 |
| chr7_-_16796309 | 0.09 |
ENSMUST00000153833.2
ENSMUST00000108492.9 |
Hif3a
|
hypoxia inducible factor 3, alpha subunit |
| chr1_-_5987617 | 0.09 |
ENSMUST00000044180.5
|
Npbwr1
|
neuropeptides B/W receptor 1 |
| chr7_+_126528016 | 0.09 |
ENSMUST00000032924.6
|
Kctd13
|
potassium channel tetramerisation domain containing 13 |
| chr10_-_127099183 | 0.09 |
ENSMUST00000099172.5
|
Kif5a
|
kinesin family member 5A |
| chr2_+_157401998 | 0.09 |
ENSMUST00000153739.9
ENSMUST00000173595.2 ENSMUST00000109526.2 ENSMUST00000173839.2 ENSMUST00000173041.8 ENSMUST00000173793.8 ENSMUST00000172487.2 ENSMUST00000088484.6 |
Nnat
|
neuronatin |
| chr15_+_101026403 | 0.09 |
ENSMUST00000000542.14
ENSMUST00000144229.8 |
Acvrl1
|
activin A receptor, type II-like 1 |
| chr1_+_153775682 | 0.09 |
ENSMUST00000086199.12
|
Glul
|
glutamate-ammonia ligase (glutamine synthetase) |
| chr4_-_46536088 | 0.09 |
ENSMUST00000102924.3
ENSMUST00000046897.13 |
Trim14
|
tripartite motif-containing 14 |
| chr11_+_53410697 | 0.08 |
ENSMUST00000120878.9
ENSMUST00000147912.2 |
Septin8
|
septin 8 |
| chr5_+_150119860 | 0.08 |
ENSMUST00000202600.4
|
Fry
|
FRY microtubule binding protein |
| chr19_-_5560473 | 0.08 |
ENSMUST00000237111.2
ENSMUST00000070172.6 |
Snx32
|
sorting nexin 32 |
| chr7_+_5054514 | 0.08 |
ENSMUST00000069324.7
|
Zfp580
|
zinc finger protein 580 |
| chr6_-_42669963 | 0.08 |
ENSMUST00000045140.5
|
Tcaf1
|
TRPM8 channel-associated factor 1 |
| chr12_-_111638722 | 0.08 |
ENSMUST00000001304.9
|
Ckb
|
creatine kinase, brain |
| chr5_-_116162415 | 0.08 |
ENSMUST00000031486.14
ENSMUST00000111999.8 |
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr15_+_100768806 | 0.08 |
ENSMUST00000201549.4
ENSMUST00000108908.6 |
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha |
| chr4_-_149569659 | 0.08 |
ENSMUST00000119921.8
|
Nmnat1
|
nicotinamide nucleotide adenylyltransferase 1 |
| chr9_+_100525637 | 0.08 |
ENSMUST00000041418.13
|
Stag1
|
stromal antigen 1 |
| chr6_+_90346547 | 0.08 |
ENSMUST00000045740.8
ENSMUST00000203493.2 |
Zxdc
|
ZXD family zinc finger C |
| chr9_+_124195807 | 0.08 |
ENSMUST00000239563.2
|
Ppp2r3d
|
protein phosphatase 2 (formerly 2A), regulatory subunit B'', delta |
| chr2_+_156681991 | 0.08 |
ENSMUST00000073352.10
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chrX_+_98086187 | 0.08 |
ENSMUST00000036606.14
|
Stard8
|
START domain containing 8 |
| chr15_-_28025920 | 0.07 |
ENSMUST00000090247.7
|
Trio
|
triple functional domain (PTPRF interacting) |
| chr4_-_155859014 | 0.07 |
ENSMUST00000042196.4
|
Vwa1
|
von Willebrand factor A domain containing 1 |
| chr11_+_113539995 | 0.07 |
ENSMUST00000018805.15
|
Cog1
|
component of oligomeric golgi complex 1 |
| chr10_+_100323420 | 0.07 |
ENSMUST00000220346.2
|
Cep290
|
centrosomal protein 290 |
| chr9_+_31191820 | 0.07 |
ENSMUST00000117389.8
ENSMUST00000215499.2 |
Prdm10
|
PR domain containing 10 |
| chr3_-_84387700 | 0.07 |
ENSMUST00000194027.2
ENSMUST00000107689.7 |
Fhdc1
|
FH2 domain containing 1 |
| chr6_-_37419030 | 0.07 |
ENSMUST00000041093.6
|
Creb3l2
|
cAMP responsive element binding protein 3-like 2 |
| chr7_+_44667377 | 0.07 |
ENSMUST00000044111.10
|
Rras
|
related RAS viral (r-ras) oncogene |
| chr8_+_84793453 | 0.07 |
ENSMUST00000211046.2
ENSMUST00000005600.6 |
Rfx1
|
regulatory factor X, 1 (influences HLA class II expression) |
| chr12_+_113061819 | 0.07 |
ENSMUST00000109727.9
ENSMUST00000009099.13 ENSMUST00000109723.8 ENSMUST00000109726.8 ENSMUST00000069690.5 |
Mta1
|
metastasis associated 1 |
| chr1_+_128031055 | 0.07 |
ENSMUST00000188381.7
ENSMUST00000187900.7 ENSMUST00000036288.11 |
R3hdm1
|
R3H domain containing 1 |
| chr17_-_17845293 | 0.07 |
ENSMUST00000041047.4
|
Lnpep
|
leucyl/cystinyl aminopeptidase |
| chr7_-_28465870 | 0.07 |
ENSMUST00000085851.12
ENSMUST00000032815.11 |
Nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, beta |
| chr7_+_100142544 | 0.07 |
ENSMUST00000126534.8
ENSMUST00000207748.2 |
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr13_+_35925296 | 0.07 |
ENSMUST00000163595.3
|
Cdyl
|
chromodomain protein, Y chromosome-like |
| chr4_-_43040278 | 0.07 |
ENSMUST00000107958.8
ENSMUST00000107959.8 ENSMUST00000152846.8 |
Fam214b
|
family with sequence similarity 214, member B |
| chr5_+_143608194 | 0.07 |
ENSMUST00000116456.10
|
Cyth3
|
cytohesin 3 |
| chr11_-_113574981 | 0.07 |
ENSMUST00000120194.2
|
Fam104a
|
family with sequence similarity 104, member A |
| chr19_-_32365862 | 0.07 |
ENSMUST00000099514.10
|
Sgms1
|
sphingomyelin synthase 1 |
| chr2_-_5680801 | 0.07 |
ENSMUST00000114987.4
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr1_+_87254729 | 0.07 |
ENSMUST00000172794.8
ENSMUST00000164992.9 ENSMUST00000173173.8 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr5_+_135093056 | 0.07 |
ENSMUST00000071263.7
|
Dnajc30
|
DnaJ heat shock protein family (Hsp40) member C30 |
| chr7_-_84339156 | 0.07 |
ENSMUST00000209117.2
ENSMUST00000207975.2 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr7_+_127111148 | 0.07 |
ENSMUST00000188124.7
ENSMUST00000189136.7 ENSMUST00000098025.11 |
Srcap
|
Snf2-related CREBBP activator protein |
| chr7_-_44741622 | 0.07 |
ENSMUST00000210469.2
ENSMUST00000211352.2 ENSMUST00000019683.11 |
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr15_+_32244947 | 0.07 |
ENSMUST00000067458.7
|
Sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr10_-_80634293 | 0.07 |
ENSMUST00000218209.2
ENSMUST00000036805.7 |
Plekhj1
|
pleckstrin homology domain containing, family J member 1 |
| chr9_-_100453102 | 0.07 |
ENSMUST00000093792.4
|
Slc35g2
|
solute carrier family 35, member G2 |
| chr16_+_37688744 | 0.07 |
ENSMUST00000078717.7
|
Lrrc58
|
leucine rich repeat containing 58 |
| chr14_+_30673334 | 0.07 |
ENSMUST00000226551.2
ENSMUST00000228328.2 |
Nek4
|
NIMA (never in mitosis gene a)-related expressed kinase 4 |
| chr2_+_156681927 | 0.07 |
ENSMUST00000081335.13
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr1_-_24139263 | 0.07 |
ENSMUST00000187369.7
ENSMUST00000187752.7 ENSMUST00000186999.7 |
Fam135a
|
family with sequence similarity 135, member A |
| chr11_+_69214895 | 0.06 |
ENSMUST00000060956.13
|
Trappc1
|
trafficking protein particle complex 1 |
| chr12_+_59113659 | 0.06 |
ENSMUST00000021381.6
|
Pnn
|
pinin |
| chr11_+_69214971 | 0.06 |
ENSMUST00000108662.2
|
Trappc1
|
trafficking protein particle complex 1 |
| chr9_-_32452885 | 0.06 |
ENSMUST00000016231.14
|
Fli1
|
Friend leukemia integration 1 |
| chr1_-_52856801 | 0.06 |
ENSMUST00000027271.9
|
Inpp1
|
inositol polyphosphate-1-phosphatase |
| chr18_-_10610332 | 0.06 |
ENSMUST00000025142.13
|
Esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr9_+_21914083 | 0.06 |
ENSMUST00000216344.2
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr5_+_149335214 | 0.06 |
ENSMUST00000093110.12
|
Medag
|
mesenteric estrogen dependent adipogenesis |
| chr4_-_120874376 | 0.06 |
ENSMUST00000043200.8
|
Smap2
|
small ArfGAP 2 |
| chr5_+_139186386 | 0.06 |
ENSMUST00000058716.14
ENSMUST00000110883.9 ENSMUST00000110884.9 ENSMUST00000100517.10 ENSMUST00000127045.8 ENSMUST00000143562.8 ENSMUST00000078690.13 ENSMUST00000146715.8 |
Sun1
|
Sad1 and UNC84 domain containing 1 |
| chr13_+_54769611 | 0.06 |
ENSMUST00000026991.16
ENSMUST00000137413.8 ENSMUST00000135232.8 ENSMUST00000124752.2 |
Faf2
|
Fas associated factor family member 2 |
| chr10_-_79602764 | 0.06 |
ENSMUST00000047203.9
|
Rnf126
|
ring finger protein 126 |
| chr6_-_124888643 | 0.06 |
ENSMUST00000032217.2
|
Lag3
|
lymphocyte-activation gene 3 |
| chr9_-_99022107 | 0.06 |
ENSMUST00000035037.14
|
Pik3cb
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta |
| chr9_-_114673158 | 0.06 |
ENSMUST00000047013.4
|
Cmtm8
|
CKLF-like MARVEL transmembrane domain containing 8 |
| chr12_-_72117865 | 0.06 |
ENSMUST00000050649.6
|
Gpr135
|
G protein-coupled receptor 135 |
| chr12_-_4088905 | 0.06 |
ENSMUST00000111178.2
|
Efr3b
|
EFR3 homolog B |
| chrX_+_18028814 | 0.06 |
ENSMUST00000044484.13
ENSMUST00000052368.9 |
Kdm6a
|
lysine (K)-specific demethylase 6A |
| chr11_-_94440025 | 0.06 |
ENSMUST00000040487.4
|
Rsad1
|
radical S-adenosyl methionine domain containing 1 |
| chr11_-_103303480 | 0.06 |
ENSMUST00000041272.10
|
Plekhm1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr1_+_87254719 | 0.06 |
ENSMUST00000027475.15
|
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr10_-_127098932 | 0.06 |
ENSMUST00000217895.2
|
Kif5a
|
kinesin family member 5A |
| chr2_-_39080701 | 0.06 |
ENSMUST00000142872.2
ENSMUST00000038874.12 |
Scai
|
suppressor of cancer cell invasion |
| chr8_-_95328260 | 0.05 |
ENSMUST00000212765.2
ENSMUST00000213008.2 ENSMUST00000212159.2 ENSMUST00000213022.2 ENSMUST00000212258.2 ENSMUST00000212788.2 ENSMUST00000212791.2 |
Psme3ip1
|
proteasome activator subunit 3 interacting protein 1 |
| chr19_-_7183596 | 0.05 |
ENSMUST00000123594.8
|
Otub1
|
OTU domain, ubiquitin aldehyde binding 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb7a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0002658 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.1 | 0.2 | GO:0061184 | positive regulation of dermatome development(GO:0061184) renal vesicle induction(GO:0072034) negative regulation of male gonad development(GO:2000019) |
| 0.1 | 0.3 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 0.4 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 0.2 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.0 | 0.5 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.2 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0002501 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.0 | 0.2 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.1 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.1 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.1 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.1 | GO:0046061 | dGTP catabolic process(GO:0006203) dATP catabolic process(GO:0046061) |
| 0.0 | 0.1 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.0 | 0.5 | GO:0045060 | negative thymic T cell selection(GO:0045060) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:1900062 | regulation of replicative cell aging(GO:1900062) |
| 0.0 | 0.3 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.1 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.2 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.0 | 0.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.0 | GO:0034118 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.0 | GO:0097045 | activation of blood coagulation via clotting cascade(GO:0002543) phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.0 | 0.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.3 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.2 | GO:0090500 | endocardial cushion to mesenchymal transition(GO:0090500) |
| 0.0 | 0.0 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.0 | 0.1 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.2 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.0 | GO:0033602 | negative regulation of dopamine secretion(GO:0033602) |
| 0.0 | 0.0 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002945 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.1 | 0.3 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.3 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.0 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.0 | 0.4 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.0 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.0 | 0.1 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.0 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.0 | 0.1 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.1 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |