Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
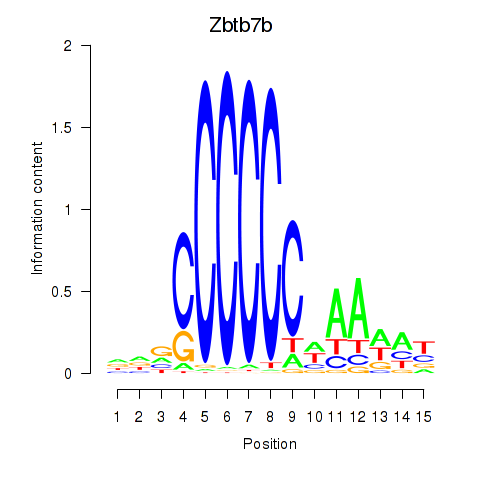
Results for Zbtb7b
Z-value: 0.59
Transcription factors associated with Zbtb7b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb7b
|
ENSMUSG00000028042.16 | zinc finger and BTB domain containing 7B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb7b | mm39_v1_chr3_-_89300599_89300685 | 0.99 | 1.1e-03 | Click! |
Activity profile of Zbtb7b motif
Sorted Z-values of Zbtb7b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_5023552 | 0.55 |
ENSMUST00000208728.2
ENSMUST00000085427.6 |
Ccdc106
Zfp865
|
coiled-coil domain containing 106 zinc finger protein 865 |
| chr17_-_57289121 | 0.49 |
ENSMUST00000056113.5
|
Acer1
|
alkaline ceramidase 1 |
| chrX_-_100838004 | 0.47 |
ENSMUST00000147742.9
|
Gm4779
|
predicted gene 4779 |
| chr12_-_99529767 | 0.44 |
ENSMUST00000176928.3
ENSMUST00000223484.2 |
Foxn3
|
forkhead box N3 |
| chr8_-_106198112 | 0.42 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr7_+_5023375 | 0.35 |
ENSMUST00000076251.7
|
Zfp865
|
zinc finger protein 865 |
| chr7_+_79675727 | 0.28 |
ENSMUST00000049680.10
|
Zfp710
|
zinc finger protein 710 |
| chr15_+_101310283 | 0.27 |
ENSMUST00000068904.9
|
Krt7
|
keratin 7 |
| chr9_-_123680726 | 0.25 |
ENSMUST00000084715.14
|
Fyco1
|
FYVE and coiled-coil domain containing 1 |
| chr6_-_72935171 | 0.24 |
ENSMUST00000114049.2
|
Tmsb10
|
thymosin, beta 10 |
| chr10_+_129153986 | 0.23 |
ENSMUST00000215503.2
|
Olfr780
|
olfactory receptor 780 |
| chr6_-_67316645 | 0.23 |
ENSMUST00000117441.8
|
Il12rb2
|
interleukin 12 receptor, beta 2 |
| chr9_-_62895197 | 0.22 |
ENSMUST00000216209.2
|
Pias1
|
protein inhibitor of activated STAT 1 |
| chrX_+_93278588 | 0.22 |
ENSMUST00000096369.10
ENSMUST00000113911.9 |
Klhl15
|
kelch-like 15 |
| chr13_-_22689551 | 0.22 |
ENSMUST00000228020.2
|
Vmn1r202
|
vomeronasal 1 receptor 202 |
| chr3_-_95125051 | 0.21 |
ENSMUST00000107204.8
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr5_-_132570710 | 0.21 |
ENSMUST00000182974.9
|
Auts2
|
autism susceptibility candidate 2 |
| chr14_-_51134930 | 0.20 |
ENSMUST00000227271.2
|
Klhl33
|
kelch-like 33 |
| chr2_+_156562956 | 0.19 |
ENSMUST00000109566.9
ENSMUST00000146412.9 ENSMUST00000177013.8 ENSMUST00000171030.9 |
Dlgap4
|
DLG associated protein 4 |
| chr7_+_28466160 | 0.18 |
ENSMUST00000122915.8
ENSMUST00000072965.5 ENSMUST00000170068.9 |
Sirt2
|
sirtuin 2 |
| chr11_+_115078299 | 0.18 |
ENSMUST00000100235.9
ENSMUST00000061450.7 |
Tmem104
|
transmembrane protein 104 |
| chr7_-_106563137 | 0.17 |
ENSMUST00000213552.2
ENSMUST00000040983.6 ENSMUST00000213651.2 |
Olfr6
|
olfactory receptor 6 |
| chr14_+_30673334 | 0.17 |
ENSMUST00000226551.2
ENSMUST00000228328.2 |
Nek4
|
NIMA (never in mitosis gene a)-related expressed kinase 4 |
| chr4_-_123421441 | 0.16 |
ENSMUST00000147228.8
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr14_+_33645539 | 0.16 |
ENSMUST00000168727.3
|
Gdf10
|
growth differentiation factor 10 |
| chr1_+_171723231 | 0.15 |
ENSMUST00000097466.3
|
Gm10521
|
predicted gene 10521 |
| chr11_-_113956996 | 0.15 |
ENSMUST00000041627.14
|
Sdk2
|
sidekick cell adhesion molecule 2 |
| chr7_+_5037117 | 0.15 |
ENSMUST00000076791.4
|
4632433K11Rik
|
RIKEN cDNA 4632433K11 gene |
| chr4_-_98712077 | 0.15 |
ENSMUST00000097964.3
|
I0C0044D17Rik
|
RIKEN cDNA I0C0044D17 gene |
| chr2_-_31031814 | 0.15 |
ENSMUST00000073879.12
ENSMUST00000100208.9 ENSMUST00000100207.9 ENSMUST00000113555.8 ENSMUST00000075326.11 ENSMUST00000113552.9 ENSMUST00000136181.8 ENSMUST00000113564.9 ENSMUST00000113562.9 ENSMUST00000113560.8 |
Fnbp1
|
formin binding protein 1 |
| chr3_-_95902949 | 0.15 |
ENSMUST00000123006.8
ENSMUST00000130043.8 |
Plekho1
|
pleckstrin homology domain containing, family O member 1 |
| chr7_-_140590605 | 0.15 |
ENSMUST00000026565.7
|
Ifitm3
|
interferon induced transmembrane protein 3 |
| chr2_+_156563284 | 0.14 |
ENSMUST00000099145.6
|
Dlgap4
|
DLG associated protein 4 |
| chr15_-_76501041 | 0.14 |
ENSMUST00000073428.7
|
Slc39a4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr6_+_108190163 | 0.13 |
ENSMUST00000203615.3
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor 1 |
| chr17_+_35455532 | 0.13 |
ENSMUST00000068261.9
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2 |
| chr7_-_28465870 | 0.13 |
ENSMUST00000085851.12
ENSMUST00000032815.11 |
Nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, beta |
| chr5_+_53748323 | 0.13 |
ENSMUST00000201883.4
|
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr2_+_32617671 | 0.13 |
ENSMUST00000113242.5
|
Sh2d3c
|
SH2 domain containing 3C |
| chr3_+_145926709 | 0.13 |
ENSMUST00000039164.4
|
Lpar3
|
lysophosphatidic acid receptor 3 |
| chr15_-_102165884 | 0.12 |
ENSMUST00000043172.15
|
Rarg
|
retinoic acid receptor, gamma |
| chr6_-_36787096 | 0.11 |
ENSMUST00000201321.2
ENSMUST00000101534.5 |
Ptn
|
pleiotrophin |
| chrX_-_139714182 | 0.11 |
ENSMUST00000044179.8
|
Tex13b
|
testis expressed 13B |
| chr9_+_56979307 | 0.11 |
ENSMUST00000169879.8
|
Sin3a
|
transcriptional regulator, SIN3A (yeast) |
| chr10_+_107998219 | 0.11 |
ENSMUST00000070663.6
|
Ppp1r12a
|
protein phosphatase 1, regulatory subunit 12A |
| chr11_+_83193495 | 0.11 |
ENSMUST00000176430.8
ENSMUST00000065692.14 ENSMUST00000142680.2 |
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr9_+_56983627 | 0.11 |
ENSMUST00000168678.8
|
Sin3a
|
transcriptional regulator, SIN3A (yeast) |
| chrX_+_7523499 | 0.11 |
ENSMUST00000033485.14
|
Prickle3
|
prickle planar cell polarity protein 3 |
| chr19_-_47525437 | 0.10 |
ENSMUST00000182808.8
ENSMUST00000182714.2 ENSMUST00000049369.16 |
Stn1
|
STN1, CST complex subunit |
| chr13_+_5911481 | 0.10 |
ENSMUST00000000080.8
|
Klf6
|
Kruppel-like factor 6 |
| chr9_+_56983679 | 0.10 |
ENSMUST00000168177.8
|
Sin3a
|
transcriptional regulator, SIN3A (yeast) |
| chr6_+_108190050 | 0.10 |
ENSMUST00000032192.9
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor 1 |
| chr16_+_8455538 | 0.10 |
ENSMUST00000023396.10
ENSMUST00000230828.2 |
Pmm2
|
phosphomannomutase 2 |
| chr2_+_156562989 | 0.09 |
ENSMUST00000000094.14
|
Dlgap4
|
DLG associated protein 4 |
| chr2_+_121120070 | 0.09 |
ENSMUST00000094639.10
|
Map1a
|
microtubule-associated protein 1 A |
| chr7_+_55443893 | 0.09 |
ENSMUST00000032627.5
|
Tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chr3_-_142101418 | 0.09 |
ENSMUST00000029941.16
ENSMUST00000058626.9 |
Pdlim5
|
PDZ and LIM domain 5 |
| chr10_-_18662526 | 0.09 |
ENSMUST00000216654.2
|
Gm4922
|
predicted gene 4922 |
| chr17_+_35454833 | 0.09 |
ENSMUST00000118384.8
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2 |
| chr7_-_80453033 | 0.08 |
ENSMUST00000167377.3
|
Iqgap1
|
IQ motif containing GTPase activating protein 1 |
| chr10_-_127016448 | 0.08 |
ENSMUST00000222911.3
ENSMUST00000095270.3 |
Slc26a10
|
solute carrier family 26, member 10 |
| chr15_-_50753792 | 0.08 |
ENSMUST00000185183.2
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr2_-_154411640 | 0.08 |
ENSMUST00000000894.6
|
E2f1
|
E2F transcription factor 1 |
| chr19_+_55730696 | 0.08 |
ENSMUST00000153888.9
ENSMUST00000127233.9 ENSMUST00000061496.17 ENSMUST00000111656.8 ENSMUST00000111657.11 |
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr12_-_98700886 | 0.08 |
ENSMUST00000085116.4
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr7_+_26853139 | 0.08 |
ENSMUST00000164093.8
|
Cyp2t4
|
cytochrome P450, family 2, subfamily t, polypeptide 4 |
| chr11_-_97909134 | 0.08 |
ENSMUST00000107561.9
|
Cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr19_-_8816530 | 0.08 |
ENSMUST00000096259.6
|
Gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr11_+_69909659 | 0.08 |
ENSMUST00000232002.2
ENSMUST00000134376.10 ENSMUST00000231221.2 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr3_+_130904000 | 0.07 |
ENSMUST00000029611.14
ENSMUST00000106341.9 ENSMUST00000066849.13 |
Lef1
|
lymphoid enhancer binding factor 1 |
| chr6_+_125169117 | 0.07 |
ENSMUST00000032485.7
|
Mrpl51
|
mitochondrial ribosomal protein L51 |
| chr13_+_112604037 | 0.07 |
ENSMUST00000183868.8
|
Il6st
|
interleukin 6 signal transducer |
| chr3_-_96501443 | 0.07 |
ENSMUST00000145001.2
ENSMUST00000091924.10 |
Polr3gl
|
polymerase (RNA) III (DNA directed) polypeptide G like |
| chr16_-_8455525 | 0.07 |
ENSMUST00000052505.10
|
Tmem186
|
transmembrane protein 186 |
| chr9_+_107458495 | 0.07 |
ENSMUST00000040059.9
|
Hyal3
|
hyaluronoglucosaminidase 3 |
| chr8_+_18645144 | 0.07 |
ENSMUST00000039412.15
|
Mcph1
|
microcephaly, primary autosomal recessive 1 |
| chr4_+_135622694 | 0.07 |
ENSMUST00000097843.9
|
Cnr2
|
cannabinoid receptor 2 (macrophage) |
| chr13_-_93328883 | 0.07 |
ENSMUST00000225868.2
|
Tent2
|
terminal nucleotidyltransferase 2 |
| chr5_+_137286535 | 0.07 |
ENSMUST00000024099.11
ENSMUST00000196208.5 ENSMUST00000085934.4 |
Ache
|
acetylcholinesterase |
| chr11_-_82655132 | 0.07 |
ENSMUST00000021040.10
ENSMUST00000100722.5 |
Cct6b
|
chaperonin containing Tcp1, subunit 6b (zeta) |
| chr2_-_117173428 | 0.07 |
ENSMUST00000102534.11
|
Rasgrp1
|
RAS guanyl releasing protein 1 |
| chr11_+_59340865 | 0.07 |
ENSMUST00000108777.10
ENSMUST00000045279.13 |
Jmjd4
|
jumonji domain containing 4 |
| chr5_+_134128543 | 0.06 |
ENSMUST00000016088.9
|
Castor2
|
cytosolic arginine sensor for mTORC1 subunit 2 |
| chr1_-_156766381 | 0.06 |
ENSMUST00000188656.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr3_+_14951264 | 0.06 |
ENSMUST00000192609.6
|
Car2
|
carbonic anhydrase 2 |
| chr13_-_93328719 | 0.06 |
ENSMUST00000048702.7
|
Tent2
|
terminal nucleotidyltransferase 2 |
| chr17_-_56416799 | 0.06 |
ENSMUST00000002908.8
ENSMUST00000238734.2 ENSMUST00000190703.7 |
Plin4
|
perilipin 4 |
| chr1_-_156766957 | 0.06 |
ENSMUST00000171292.8
ENSMUST00000063199.13 ENSMUST00000027886.14 |
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr13_-_53627110 | 0.06 |
ENSMUST00000021922.10
|
Msx2
|
msh homeobox 2 |
| chrX_+_165127688 | 0.06 |
ENSMUST00000112223.8
ENSMUST00000112224.8 ENSMUST00000112229.9 ENSMUST00000112228.8 ENSMUST00000112227.9 ENSMUST00000112226.3 |
Gpm6b
|
glycoprotein m6b |
| chr15_+_97259060 | 0.06 |
ENSMUST00000228521.2
ENSMUST00000226495.2 |
Pced1b
|
PC-esterase domain containing 1B |
| chr2_-_117173312 | 0.06 |
ENSMUST00000178884.8
|
Rasgrp1
|
RAS guanyl releasing protein 1 |
| chr7_-_126062272 | 0.06 |
ENSMUST00000032974.13
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr2_+_25346841 | 0.06 |
ENSMUST00000114265.9
ENSMUST00000102918.3 |
Clic3
|
chloride intracellular channel 3 |
| chr11_+_75404356 | 0.05 |
ENSMUST00000042808.13
ENSMUST00000118243.2 |
Scarf1
|
scavenger receptor class F, member 1 |
| chr7_-_140793990 | 0.05 |
ENSMUST00000026573.7
ENSMUST00000170841.9 |
Lmntd2
|
lamin tail domain containing 2 |
| chr8_+_84699580 | 0.05 |
ENSMUST00000005606.8
|
Prkaca
|
protein kinase, cAMP dependent, catalytic, alpha |
| chr2_-_154411765 | 0.05 |
ENSMUST00000103145.11
|
E2f1
|
E2F transcription factor 1 |
| chr5_+_110324734 | 0.05 |
ENSMUST00000139611.8
ENSMUST00000031477.9 |
Golga3
|
golgi autoantigen, golgin subfamily a, 3 |
| chr10_+_84591919 | 0.05 |
ENSMUST00000060397.13
|
Rfx4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr7_+_55443931 | 0.05 |
ENSMUST00000205796.2
|
Tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chr11_-_75345482 | 0.05 |
ENSMUST00000173320.8
|
Wdr81
|
WD repeat domain 81 |
| chr15_-_102165740 | 0.04 |
ENSMUST00000135466.2
|
Rarg
|
retinoic acid receptor, gamma |
| chr1_+_40123858 | 0.04 |
ENSMUST00000027243.13
|
Il1r2
|
interleukin 1 receptor, type II |
| chr6_-_141892517 | 0.04 |
ENSMUST00000168119.8
|
Slco1a1
|
solute carrier organic anion transporter family, member 1a1 |
| chr19_+_6887059 | 0.04 |
ENSMUST00000088257.14
ENSMUST00000116551.10 |
Trmt112
|
tRNA methyltransferase 11-2 |
| chr7_+_128289261 | 0.04 |
ENSMUST00000151237.5
|
Inpp5f
|
inositol polyphosphate-5-phosphatase F |
| chr6_-_69245427 | 0.04 |
ENSMUST00000103348.3
|
Igkv4-70
|
immunoglobulin kappa chain variable 4-70 |
| chr19_+_6887486 | 0.04 |
ENSMUST00000174786.2
|
Trmt112
|
tRNA methyltransferase 11-2 |
| chr16_+_44586085 | 0.04 |
ENSMUST00000057488.15
|
Cd200r1
|
CD200 receptor 1 |
| chrX_+_20570145 | 0.04 |
ENSMUST00000033383.3
|
Usp11
|
ubiquitin specific peptidase 11 |
| chr17_+_34124078 | 0.04 |
ENSMUST00000172817.2
|
Smim40
|
small integral membrane protein 40 |
| chr17_+_48653493 | 0.04 |
ENSMUST00000113237.4
|
Trem2
|
triggering receptor expressed on myeloid cells 2 |
| chr7_-_4525793 | 0.04 |
ENSMUST00000140424.8
|
Tnni3
|
troponin I, cardiac 3 |
| chr11_-_113600346 | 0.04 |
ENSMUST00000173655.8
ENSMUST00000100248.6 |
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr4_+_117692583 | 0.03 |
ENSMUST00000169885.8
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr9_-_20553576 | 0.03 |
ENSMUST00000155301.8
|
Fbxl12
|
F-box and leucine-rich repeat protein 12 |
| chr6_+_87019819 | 0.03 |
ENSMUST00000113657.8
ENSMUST00000113658.8 ENSMUST00000113655.8 ENSMUST00000032057.8 |
Gfpt1
|
glutamine fructose-6-phosphate transaminase 1 |
| chr18_+_65158873 | 0.03 |
ENSMUST00000226058.2
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr11_+_87959067 | 0.03 |
ENSMUST00000018521.11
|
Vezf1
|
vascular endothelial zinc finger 1 |
| chr7_-_126398343 | 0.03 |
ENSMUST00000032934.12
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr7_-_4755971 | 0.03 |
ENSMUST00000183971.8
ENSMUST00000182173.2 ENSMUST00000182738.8 ENSMUST00000182111.8 ENSMUST00000184143.8 ENSMUST00000182048.2 ENSMUST00000063324.14 |
Cox6b2
|
cytochrome c oxidase subunit 6B2 |
| chr11_-_32217547 | 0.03 |
ENSMUST00000109389.9
ENSMUST00000129010.2 ENSMUST00000020530.12 |
Nprl3
|
nitrogen permease regulator-like 3 |
| chr10_+_34173426 | 0.03 |
ENSMUST00000047935.8
|
Tspyl4
|
TSPY-like 4 |
| chr5_+_110324506 | 0.03 |
ENSMUST00000112512.8
|
Golga3
|
golgi autoantigen, golgin subfamily a, 3 |
| chr7_+_55443967 | 0.03 |
ENSMUST00000206454.2
|
Tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chr1_+_153776323 | 0.03 |
ENSMUST00000140685.4
ENSMUST00000139476.8 |
Glul
|
glutamate-ammonia ligase (glutamine synthetase) |
| chr12_-_101924407 | 0.03 |
ENSMUST00000159883.2
ENSMUST00000160251.8 ENSMUST00000161011.8 ENSMUST00000021606.12 |
Atxn3
|
ataxin 3 |
| chr7_-_142211203 | 0.03 |
ENSMUST00000097936.9
ENSMUST00000000033.12 |
Igf2
|
insulin-like growth factor 2 |
| chr6_-_113694633 | 0.03 |
ENSMUST00000204533.3
|
Ghrl
|
ghrelin |
| chr4_-_133329479 | 0.03 |
ENSMUST00000057311.4
|
Sfn
|
stratifin |
| chr1_-_156766351 | 0.03 |
ENSMUST00000189648.2
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr7_+_119393210 | 0.02 |
ENSMUST00000033218.15
ENSMUST00000106520.9 |
Rexo5
|
RNA exonuclease 5 |
| chr15_-_76259126 | 0.02 |
ENSMUST00000071119.8
|
Tssk5
|
testis-specific serine kinase 5 |
| chr6_-_69415741 | 0.02 |
ENSMUST00000103354.3
|
Igkv4-59
|
immunoglobulin kappa variable 4-59 |
| chr19_+_55730242 | 0.02 |
ENSMUST00000111662.11
ENSMUST00000041717.14 |
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr4_+_129941633 | 0.02 |
ENSMUST00000044565.15
ENSMUST00000132251.2 |
Col16a1
|
collagen, type XVI, alpha 1 |
| chr11_+_101623776 | 0.02 |
ENSMUST00000039152.14
|
Dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr7_-_30754240 | 0.02 |
ENSMUST00000206860.2
ENSMUST00000071697.11 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr17_+_56266146 | 0.02 |
ENSMUST00000086869.6
|
Yju2
|
YJU2 splicing factor |
| chr17_+_34311314 | 0.02 |
ENSMUST00000025192.8
|
H2-Oa
|
histocompatibility 2, O region alpha locus |
| chr12_-_84497718 | 0.02 |
ENSMUST00000085192.7
ENSMUST00000220491.2 |
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1 |
| chr7_-_30754223 | 0.02 |
ENSMUST00000206012.2
ENSMUST00000108110.5 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr17_+_34544632 | 0.02 |
ENSMUST00000050325.10
|
H2-Eb2
|
histocompatibility 2, class II antigen E beta2 |
| chr17_+_56266099 | 0.02 |
ENSMUST00000233661.2
|
Yju2
|
YJU2 splicing factor |
| chr7_-_130964469 | 0.02 |
ENSMUST00000059438.11
|
2310057M21Rik
|
RIKEN cDNA 2310057M21 gene |
| chr14_-_78326435 | 0.02 |
ENSMUST00000118785.3
ENSMUST00000066437.5 |
Fam216b
|
family with sequence similarity 216, member B |
| chr2_-_118377500 | 0.02 |
ENSMUST00000125860.3
|
Bmf
|
BCL2 modifying factor |
| chr15_+_95688763 | 0.02 |
ENSMUST00000227791.2
|
Ano6
|
anoctamin 6 |
| chr7_+_6733684 | 0.01 |
ENSMUST00000197117.5
|
Usp29
|
ubiquitin specific peptidase 29 |
| chr7_+_25016492 | 0.01 |
ENSMUST00000128119.2
|
Megf8
|
multiple EGF-like-domains 8 |
| chr2_-_160155536 | 0.01 |
ENSMUST00000109475.3
|
Gm826
|
predicted gene 826 |
| chr12_-_111874489 | 0.01 |
ENSMUST00000054815.15
|
Ppp1r13b
|
protein phosphatase 1, regulatory subunit 13B |
| chr11_+_20581952 | 0.01 |
ENSMUST00000109586.3
|
Sertad2
|
SERTA domain containing 2 |
| chr9_+_113570506 | 0.01 |
ENSMUST00000213663.2
|
Clasp2
|
CLIP associating protein 2 |
| chr3_+_52175757 | 0.01 |
ENSMUST00000053764.7
|
Foxo1
|
forkhead box O1 |
| chr18_-_34757653 | 0.01 |
ENSMUST00000003876.10
ENSMUST00000115766.8 ENSMUST00000097626.10 ENSMUST00000115765.2 |
Brd8
|
bromodomain containing 8 |
| chr12_+_113038098 | 0.01 |
ENSMUST00000012355.14
ENSMUST00000146107.8 |
Tex22
|
testis expressed gene 22 |
| chr1_-_151304191 | 0.01 |
ENSMUST00000064771.12
|
Swt1
|
SWT1 RNA endoribonuclease homolog (S. cerevisiae) |
| chr8_-_105036664 | 0.01 |
ENSMUST00000160596.8
ENSMUST00000164175.2 |
Cmtm1
|
CKLF-like MARVEL transmembrane domain containing 1 |
| chr5_-_123620632 | 0.01 |
ENSMUST00000198901.2
|
Il31
|
interleukin 31 |
| chr6_-_85879510 | 0.01 |
ENSMUST00000159755.8
|
Nat8f4
|
N-acetyltransferase 8 (GCN5-related) family member 4 |
| chr7_-_4973960 | 0.01 |
ENSMUST00000144863.8
|
Sbk3
|
SH3 domain binding kinase family, member 3 |
| chr7_-_97387145 | 0.01 |
ENSMUST00000084986.8
|
Aqp11
|
aquaporin 11 |
| chr4_-_108075119 | 0.01 |
ENSMUST00000223127.2
ENSMUST00000043793.7 ENSMUST00000106690.9 |
Zyg11a
|
zyg-11 family member A, cell cycle regulator |
| chr3_-_95125190 | 0.01 |
ENSMUST00000136139.8
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr9_+_110075133 | 0.01 |
ENSMUST00000199736.2
|
Cspg5
|
chondroitin sulfate proteoglycan 5 |
| chr15_+_85716503 | 0.01 |
ENSMUST00000146088.8
|
Ttc38
|
tetratricopeptide repeat domain 38 |
| chr1_+_75522902 | 0.01 |
ENSMUST00000124341.8
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr8_-_105036739 | 0.01 |
ENSMUST00000159039.2
|
Cmtm1
|
CKLF-like MARVEL transmembrane domain containing 1 |
| chrX_+_151016224 | 0.01 |
ENSMUST00000112588.9
ENSMUST00000082177.13 |
Kdm5c
|
lysine (K)-specific demethylase 5C |
| chr19_-_46314945 | 0.01 |
ENSMUST00000225781.2
ENSMUST00000223903.2 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr8_-_106140106 | 0.01 |
ENSMUST00000167294.8
ENSMUST00000063071.13 |
Kctd19
|
potassium channel tetramerisation domain containing 19 |
| chr15_-_3333003 | 0.01 |
ENSMUST00000165386.2
|
Ccdc152
|
coiled-coil domain containing 152 |
| chrX_+_165021919 | 0.01 |
ENSMUST00000060210.14
ENSMUST00000112233.8 |
Gpm6b
|
glycoprotein m6b |
| chr11_-_113600838 | 0.01 |
ENSMUST00000018871.8
|
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr7_+_18817767 | 0.01 |
ENSMUST00000032568.14
ENSMUST00000122999.8 ENSMUST00000108473.10 ENSMUST00000108474.2 ENSMUST00000238982.2 |
Dmpk
|
dystrophia myotonica-protein kinase |
| chr2_-_32665596 | 0.01 |
ENSMUST00000161430.8
|
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr5_+_91665474 | 0.01 |
ENSMUST00000040576.10
|
Parm1
|
prostate androgen-regulated mucin-like protein 1 |
| chr19_-_13807805 | 0.01 |
ENSMUST00000214475.2
ENSMUST00000067670.5 ENSMUST00000219674.2 ENSMUST00000216287.2 ENSMUST00000215760.2 |
Olfr1500
|
olfactory receptor 1500 |
| chr13_-_112788829 | 0.01 |
ENSMUST00000075748.7
|
Ddx4
|
DEAD box helicase 4 |
| chrX_+_8137620 | 0.01 |
ENSMUST00000033512.11
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr3_-_46402355 | 0.01 |
ENSMUST00000195537.2
ENSMUST00000166505.7 ENSMUST00000195436.2 |
Pabpc4l
|
poly(A) binding protein, cytoplasmic 4-like |
| chr19_+_55730488 | 0.01 |
ENSMUST00000111659.9
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr19_-_45800730 | 0.01 |
ENSMUST00000086993.11
|
Kcnip2
|
Kv channel-interacting protein 2 |
| chr17_+_33651864 | 0.01 |
ENSMUST00000174088.3
|
Actl9
|
actin-like 9 |
| chr11_+_82655158 | 0.01 |
ENSMUST00000056677.8
|
Zfp830
|
zinc finger protein 830 |
| chr2_+_91757594 | 0.00 |
ENSMUST00000045537.4
|
Chrm4
|
cholinergic receptor, muscarinic 4 |
| chr14_-_51134906 | 0.00 |
ENSMUST00000170855.2
|
Klhl33
|
kelch-like 33 |
| chr1_+_75213258 | 0.00 |
ENSMUST00000185654.3
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr2_+_128704867 | 0.00 |
ENSMUST00000110324.8
|
Fbln7
|
fibulin 7 |
| chr9_-_121668527 | 0.00 |
ENSMUST00000135986.9
|
Ccdc13
|
coiled-coil domain containing 13 |
| chr15_+_76152276 | 0.00 |
ENSMUST00000074173.4
|
Spatc1
|
spermatogenesis and centriole associated 1 |
| chr14_+_65835995 | 0.00 |
ENSMUST00000150897.8
|
Nuggc
|
nuclear GTPase, germinal center associated |
| chr10_-_21943978 | 0.00 |
ENSMUST00000092672.6
|
4930444G20Rik
|
RIKEN cDNA 4930444G20 gene |
| chr12_+_113038376 | 0.00 |
ENSMUST00000109729.3
|
Tex22
|
testis expressed gene 22 |
| chr11_+_104077153 | 0.00 |
ENSMUST00000107000.2
ENSMUST00000059448.8 |
Sppl2c
|
signal peptide peptidase 2C |
| chr10_-_87329513 | 0.00 |
ENSMUST00000020243.10
|
Ascl1
|
achaete-scute family bHLH transcription factor 1 |
| chr17_-_33136021 | 0.00 |
ENSMUST00000054174.9
|
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr2_+_164404499 | 0.00 |
ENSMUST00000017867.10
ENSMUST00000109344.9 ENSMUST00000109345.9 |
Wfdc2
|
WAP four-disulfide core domain 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb7b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.1 | 0.3 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 0.2 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.1 | 0.2 | GO:1900062 | regulation of replicative cell aging(GO:1900062) |
| 0.1 | 0.2 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.2 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.2 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.2 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.1 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.0 | 0.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.1 | GO:1901297 | positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0070104 | negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0045212 | neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.1 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.0 | 0.1 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.1 | GO:0061152 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) |
| 0.0 | 0.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.5 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:2000878 | positive regulation of cellular pH reduction(GO:0032849) dipeptide transmembrane transport(GO:0035442) regulation of oligopeptide transport(GO:0090088) regulation of dipeptide transport(GO:0090089) positive regulation of oligopeptide transport(GO:2000878) positive regulation of dipeptide transport(GO:2000880) regulation of dipeptide transmembrane transport(GO:2001148) positive regulation of dipeptide transmembrane transport(GO:2001150) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.2 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.0 | GO:0002582 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) detection of peptidoglycan(GO:0032499) |
| 0.0 | 0.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.0 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.0 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.1 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0002586 | regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.2 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.1 | GO:0004615 | phosphomannomutase activity(GO:0004615) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.0 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |