Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
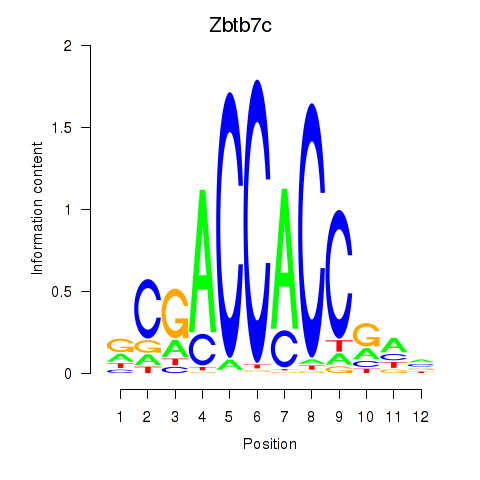
Results for Zbtb7c
Z-value: 0.35
Transcription factors associated with Zbtb7c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb7c
|
ENSMUSG00000044646.16 | zinc finger and BTB domain containing 7C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb7c | mm39_v1_chr18_+_76192529_76192571 | -0.43 | 4.7e-01 | Click! |
Activity profile of Zbtb7c motif
Sorted Z-values of Zbtb7c motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_6863269 | 0.25 |
ENSMUST00000171311.8
ENSMUST00000160937.9 |
Dlx6
|
distal-less homeobox 6 |
| chr12_-_76756772 | 0.24 |
ENSMUST00000166101.2
|
Sptb
|
spectrin beta, erythrocytic |
| chrX_-_71318353 | 0.21 |
ENSMUST00000064780.4
|
Gabre
|
gamma-aminobutyric acid (GABA) A receptor, subunit epsilon |
| chr11_+_68322945 | 0.20 |
ENSMUST00000021283.8
|
Pik3r5
|
phosphoinositide-3-kinase regulatory subunit 5 |
| chr8_+_91635192 | 0.19 |
ENSMUST00000211403.2
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr4_-_123558516 | 0.14 |
ENSMUST00000147030.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr7_-_15781838 | 0.14 |
ENSMUST00000210781.2
|
Bicra
|
BRD4 interacting chromatin remodeling complex associated protein |
| chr2_+_157401998 | 0.13 |
ENSMUST00000153739.9
ENSMUST00000173595.2 ENSMUST00000109526.2 ENSMUST00000173839.2 ENSMUST00000173041.8 ENSMUST00000173793.8 ENSMUST00000172487.2 ENSMUST00000088484.6 |
Nnat
|
neuronatin |
| chr18_-_46345661 | 0.13 |
ENSMUST00000037011.6
|
Trim36
|
tripartite motif-containing 36 |
| chrX_+_7439839 | 0.12 |
ENSMUST00000144719.9
ENSMUST00000234896.2 |
Flicr
Foxp3
|
Foxp3 regulating long intergenic noncoding RNA forkhead box P3 |
| chr6_-_124888643 | 0.12 |
ENSMUST00000032217.2
|
Lag3
|
lymphocyte-activation gene 3 |
| chr6_+_136931519 | 0.11 |
ENSMUST00000137768.2
|
Pde6h
|
phosphodiesterase 6H, cGMP-specific, cone, gamma |
| chr9_+_67747668 | 0.11 |
ENSMUST00000077879.7
|
Vps13c
|
vacuolar protein sorting 13C |
| chr11_+_70453666 | 0.11 |
ENSMUST00000072237.13
ENSMUST00000072873.14 |
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr9_+_31191820 | 0.11 |
ENSMUST00000117389.8
ENSMUST00000215499.2 |
Prdm10
|
PR domain containing 10 |
| chr11_+_75422516 | 0.11 |
ENSMUST00000149727.8
ENSMUST00000108433.8 ENSMUST00000042561.14 ENSMUST00000143035.8 |
Slc43a2
|
solute carrier family 43, member 2 |
| chr11_+_70453724 | 0.11 |
ENSMUST00000102559.11
|
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr11_+_75422925 | 0.10 |
ENSMUST00000169547.9
|
Slc43a2
|
solute carrier family 43, member 2 |
| chr10_-_80382611 | 0.10 |
ENSMUST00000183233.9
ENSMUST00000182604.8 |
Rexo1
|
REX1, RNA exonuclease 1 |
| chr11_-_70578775 | 0.10 |
ENSMUST00000036299.14
ENSMUST00000119120.2 ENSMUST00000100933.10 |
Camta2
|
calmodulin binding transcription activator 2 |
| chr15_+_78783867 | 0.09 |
ENSMUST00000134703.8
ENSMUST00000061239.14 ENSMUST00000109698.9 |
Gm49510
Sh3bp1
|
predicted gene, 49510 SH3-domain binding protein 1 |
| chr16_+_43993599 | 0.09 |
ENSMUST00000119746.8
ENSMUST00000088356.10 ENSMUST00000169582.3 |
Usf3
|
upstream transcription factor family member 3 |
| chr10_-_43880353 | 0.09 |
ENSMUST00000020017.14
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr9_+_66257747 | 0.09 |
ENSMUST00000042824.13
|
Herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr11_+_69656725 | 0.09 |
ENSMUST00000108640.8
ENSMUST00000108639.8 |
Zbtb4
|
zinc finger and BTB domain containing 4 |
| chr15_+_78784043 | 0.09 |
ENSMUST00000001226.11
|
Sh3bp1
|
SH3-domain binding protein 1 |
| chr11_+_70453806 | 0.08 |
ENSMUST00000079244.12
ENSMUST00000102558.11 |
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr11_+_53410697 | 0.08 |
ENSMUST00000120878.9
ENSMUST00000147912.2 |
Septin8
|
septin 8 |
| chr11_+_69656797 | 0.08 |
ENSMUST00000108642.8
ENSMUST00000156932.8 |
Zbtb4
|
zinc finger and BTB domain containing 4 |
| chr16_-_48814437 | 0.08 |
ENSMUST00000121869.8
|
Dzip3
|
DAZ interacting protein 3, zinc finger |
| chr10_-_127098932 | 0.08 |
ENSMUST00000217895.2
|
Kif5a
|
kinesin family member 5A |
| chr11_-_70578744 | 0.07 |
ENSMUST00000108545.9
ENSMUST00000120261.8 |
Camta2
|
calmodulin binding transcription activator 2 |
| chr7_+_27879650 | 0.06 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr3_-_142101418 | 0.06 |
ENSMUST00000029941.16
ENSMUST00000058626.9 |
Pdlim5
|
PDZ and LIM domain 5 |
| chr11_-_70578905 | 0.06 |
ENSMUST00000108544.8
|
Camta2
|
calmodulin binding transcription activator 2 |
| chr16_-_48814294 | 0.06 |
ENSMUST00000114516.8
|
Dzip3
|
DAZ interacting protein 3, zinc finger |
| chr10_-_127099183 | 0.06 |
ENSMUST00000099172.5
|
Kif5a
|
kinesin family member 5A |
| chr7_-_28135616 | 0.06 |
ENSMUST00000208199.2
|
Samd4b
|
sterile alpha motif domain containing 4B |
| chr11_+_53410552 | 0.06 |
ENSMUST00000108987.8
ENSMUST00000121334.8 ENSMUST00000117061.8 |
Septin8
|
septin 8 |
| chr11_+_120612278 | 0.05 |
ENSMUST00000018156.12
|
Rac3
|
Rac family small GTPase 3 |
| chr15_+_81469538 | 0.05 |
ENSMUST00000068387.11
|
Ep300
|
E1A binding protein p300 |
| chr7_-_34755985 | 0.05 |
ENSMUST00000130491.3
|
Cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr13_+_38010879 | 0.05 |
ENSMUST00000149745.8
|
Rreb1
|
ras responsive element binding protein 1 |
| chrX_-_47297436 | 0.05 |
ENSMUST00000037960.11
|
Zdhhc9
|
zinc finger, DHHC domain containing 9 |
| chr8_+_72993913 | 0.05 |
ENSMUST00000145213.8
|
Ap1m1
|
adaptor-related protein complex AP-1, mu subunit 1 |
| chr14_-_54924062 | 0.05 |
ENSMUST00000147714.8
|
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr16_+_4867876 | 0.05 |
ENSMUST00000230703.2
ENSMUST00000052449.6 |
Ubn1
|
ubinuclein 1 |
| chr1_-_36484328 | 0.04 |
ENSMUST00000125304.8
ENSMUST00000115011.8 |
Lman2l
|
lectin, mannose-binding 2-like |
| chr7_-_79492091 | 0.04 |
ENSMUST00000049004.8
|
Anpep
|
alanyl (membrane) aminopeptidase |
| chr18_+_34910064 | 0.04 |
ENSMUST00000043775.9
ENSMUST00000224715.2 |
Kdm3b
|
KDM3B lysine (K)-specific demethylase 3B |
| chr10_+_74896383 | 0.04 |
ENSMUST00000164107.3
|
Bcr
|
BCR activator of RhoGEF and GTPase |
| chr19_-_5323092 | 0.04 |
ENSMUST00000237463.2
ENSMUST00000025786.9 |
Pacs1
|
phosphofurin acidic cluster sorting protein 1 |
| chr3_-_96634769 | 0.04 |
ENSMUST00000141377.8
ENSMUST00000125183.2 |
Polr3c
|
polymerase (RNA) III (DNA directed) polypeptide C |
| chr11_-_70120503 | 0.03 |
ENSMUST00000153449.2
ENSMUST00000000326.12 |
Bcl6b
|
B cell CLL/lymphoma 6, member B |
| chr19_-_46306506 | 0.03 |
ENSMUST00000224556.2
|
Psd
|
pleckstrin and Sec7 domain containing |
| chr7_+_5059703 | 0.03 |
ENSMUST00000208042.2
ENSMUST00000207974.2 |
Ccdc106
|
coiled-coil domain containing 106 |
| chr12_-_4088905 | 0.03 |
ENSMUST00000111178.2
|
Efr3b
|
EFR3 homolog B |
| chr11_+_120612369 | 0.03 |
ENSMUST00000142229.2
|
Rac3
|
Rac family small GTPase 3 |
| chr11_+_79980210 | 0.03 |
ENSMUST00000017694.7
ENSMUST00000108239.7 |
Atad5
|
ATPase family, AAA domain containing 5 |
| chr11_+_101066867 | 0.03 |
ENSMUST00000103109.4
|
Cntnap1
|
contactin associated protein-like 1 |
| chr7_+_135139542 | 0.03 |
ENSMUST00000073961.8
|
Ptpre
|
protein tyrosine phosphatase, receptor type, E |
| chr8_+_33876353 | 0.03 |
ENSMUST00000070340.6
ENSMUST00000078058.5 |
Purg
|
purine-rich element binding protein G |
| chr14_-_54923517 | 0.03 |
ENSMUST00000125265.2
|
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr1_+_55127110 | 0.03 |
ENSMUST00000075242.7
|
Hspe1
|
heat shock protein 1 (chaperonin 10) |
| chr1_-_171188302 | 0.03 |
ENSMUST00000061878.5
|
Klhdc9
|
kelch domain containing 9 |
| chr7_+_79939747 | 0.03 |
ENSMUST00000205864.2
|
Vps33b
|
vacuolar protein sorting 33B |
| chr5_-_148865429 | 0.03 |
ENSMUST00000149169.3
ENSMUST00000047257.15 |
Katnal1
|
katanin p60 subunit A-like 1 |
| chr5_+_108416763 | 0.03 |
ENSMUST00000031190.5
|
Dr1
|
down-regulator of transcription 1 |
| chr11_+_21189277 | 0.03 |
ENSMUST00000109578.8
ENSMUST00000006221.14 |
Vps54
|
VPS54 GARP complex subunit |
| chr15_+_76131020 | 0.03 |
ENSMUST00000229380.2
|
Grina
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr5_-_137784912 | 0.03 |
ENSMUST00000031740.16
|
Mepce
|
methylphosphate capping enzyme |
| chr2_-_32271833 | 0.02 |
ENSMUST00000146423.2
|
1110008P14Rik
|
RIKEN cDNA 1110008P14 gene |
| chr1_+_34840785 | 0.02 |
ENSMUST00000047664.16
ENSMUST00000211073.2 |
Arhgef4
SMIM39
|
Rho guanine nucleotide exchange factor (GEF) 4 novel protein |
| chr8_+_4216556 | 0.02 |
ENSMUST00000239400.2
ENSMUST00000177053.8 ENSMUST00000176149.9 ENSMUST00000176072.9 ENSMUST00000176825.3 |
Evi5l
|
ecotropic viral integration site 5 like |
| chr3_-_96634880 | 0.02 |
ENSMUST00000029741.9
|
Polr3c
|
polymerase (RNA) III (DNA directed) polypeptide C |
| chr1_-_106641940 | 0.02 |
ENSMUST00000112751.2
|
Bcl2
|
B cell leukemia/lymphoma 2 |
| chr8_+_72993862 | 0.02 |
ENSMUST00000003117.15
ENSMUST00000212841.2 |
Ap1m1
|
adaptor-related protein complex AP-1, mu subunit 1 |
| chr4_-_126150066 | 0.02 |
ENSMUST00000122129.8
ENSMUST00000061143.15 ENSMUST00000106132.3 |
Map7d1
|
MAP7 domain containing 1 |
| chr1_-_162305573 | 0.02 |
ENSMUST00000086074.12
ENSMUST00000070330.14 |
Dnm3
|
dynamin 3 |
| chr12_+_102094977 | 0.02 |
ENSMUST00000159329.8
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr8_+_84699580 | 0.01 |
ENSMUST00000005606.8
|
Prkaca
|
protein kinase, cAMP dependent, catalytic, alpha |
| chr10_+_84938452 | 0.01 |
ENSMUST00000095383.6
|
Tmem263
|
transmembrane protein 263 |
| chr11_+_104122291 | 0.01 |
ENSMUST00000145227.8
|
Mapt
|
microtubule-associated protein tau |
| chr12_+_31315270 | 0.01 |
ENSMUST00000002979.16
ENSMUST00000239496.2 ENSMUST00000170495.3 |
Lamb1
|
laminin B1 |
| chrX_+_151016224 | 0.01 |
ENSMUST00000112588.9
ENSMUST00000082177.13 |
Kdm5c
|
lysine (K)-specific demethylase 5C |
| chr12_-_110807330 | 0.01 |
ENSMUST00000177224.2
ENSMUST00000084974.11 ENSMUST00000070565.15 |
Mok
|
MOK protein kinase |
| chr18_-_12369351 | 0.01 |
ENSMUST00000025279.6
|
Npc1
|
NPC intracellular cholesterol transporter 1 |
| chr3_-_96634752 | 0.01 |
ENSMUST00000154679.8
|
Polr3c
|
polymerase (RNA) III (DNA directed) polypeptide C |
| chr5_+_30869193 | 0.01 |
ENSMUST00000088081.11
ENSMUST00000101442.4 |
Dpysl5
|
dihydropyrimidinase-like 5 |
| chr2_+_158636727 | 0.01 |
ENSMUST00000029186.14
ENSMUST00000109478.9 ENSMUST00000156893.2 |
Dhx35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr19_-_21630143 | 0.01 |
ENSMUST00000179768.8
ENSMUST00000178523.2 ENSMUST00000038830.10 |
1110059E24Rik
|
RIKEN cDNA 1110059E24 gene |
| chr2_+_119181703 | 0.01 |
ENSMUST00000028780.4
|
Chac1
|
ChaC, cation transport regulator 1 |
| chr7_+_44667377 | 0.01 |
ENSMUST00000044111.10
|
Rras
|
related RAS viral (r-ras) oncogene |
| chr16_+_17379749 | 0.01 |
ENSMUST00000171002.10
ENSMUST00000023441.11 |
P2rx6
|
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chr7_+_57240250 | 0.00 |
ENSMUST00000196198.5
|
Gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 3 |
| chr7_+_57240894 | 0.00 |
ENSMUST00000039697.14
|
Gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 3 |
| chr16_-_4867703 | 0.00 |
ENSMUST00000115844.3
ENSMUST00000023189.15 |
Glyr1
|
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr11_+_75422953 | 0.00 |
ENSMUST00000127226.3
|
Slc43a2
|
solute carrier family 43, member 2 |
| chr8_-_35432783 | 0.00 |
ENSMUST00000033929.6
|
Tnks
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr7_+_90125890 | 0.00 |
ENSMUST00000208919.2
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr12_+_51640097 | 0.00 |
ENSMUST00000164782.10
ENSMUST00000085412.7 |
Coch
|
cochlin |
| chr11_-_99176086 | 0.00 |
ENSMUST00000017255.4
|
Krt24
|
keratin 24 |
| chr2_+_160573178 | 0.00 |
ENSMUST00000103115.8
|
Plcg1
|
phospholipase C, gamma 1 |
| chr11_-_102338473 | 0.00 |
ENSMUST00000049057.5
|
Fam171a2
|
family with sequence similarity 171, member A2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb7c
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002660 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.0 | 0.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.1 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.1 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0014737 | positive regulation of muscle atrophy(GO:0014737) |
| 0.0 | 0.3 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0010428 | methyl-CpNpG binding(GO:0010428) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |