Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
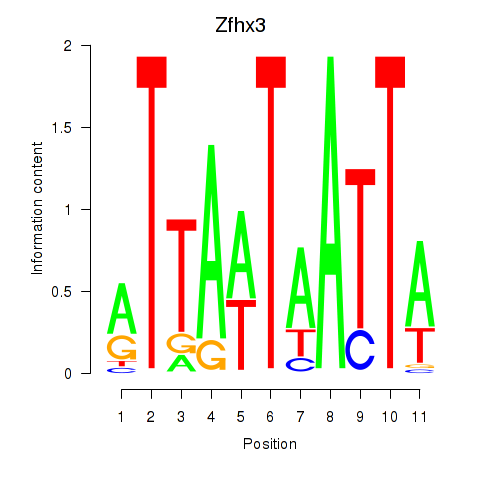
Results for Zfhx3
Z-value: 0.72
Transcription factors associated with Zfhx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfhx3
|
ENSMUSG00000038872.11 | zinc finger homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfhx3 | mm39_v1_chr8_+_109441276_109441276 | -0.70 | 1.9e-01 | Click! |
Activity profile of Zfhx3 motif
Sorted Z-values of Zfhx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_154721288 | 0.71 |
ENSMUST00000030902.13
ENSMUST00000105637.8 ENSMUST00000070313.14 ENSMUST00000105636.8 ENSMUST00000105638.9 ENSMUST00000097759.9 ENSMUST00000124771.2 |
Prdm16
|
PR domain containing 16 |
| chrM_+_7779 | 0.47 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr10_+_79650496 | 0.44 |
ENSMUST00000218857.2
ENSMUST00000220365.2 |
Palm
|
paralemmin |
| chrM_+_9459 | 0.41 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr14_+_26722319 | 0.38 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr11_+_78356523 | 0.36 |
ENSMUST00000001126.4
|
Slc46a1
|
solute carrier family 46, member 1 |
| chrM_+_7758 | 0.35 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chrM_+_8603 | 0.28 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chrM_+_2743 | 0.27 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr1_+_179788037 | 0.27 |
ENSMUST00000097453.9
ENSMUST00000111117.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr18_-_66155651 | 0.24 |
ENSMUST00000143990.2
|
Lman1
|
lectin, mannose-binding, 1 |
| chrM_+_7006 | 0.20 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr1_+_179788675 | 0.20 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chrM_+_9870 | 0.19 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr3_-_130524024 | 0.18 |
ENSMUST00000079085.11
|
Rpl34
|
ribosomal protein L34 |
| chr11_+_3933636 | 0.16 |
ENSMUST00000078757.8
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr6_-_86742847 | 0.16 |
ENSMUST00000113675.8
|
Anxa4
|
annexin A4 |
| chr4_+_98919183 | 0.16 |
ENSMUST00000030280.7
|
Angptl3
|
angiopoietin-like 3 |
| chr6_-_86742789 | 0.16 |
ENSMUST00000123732.4
|
Anxa4
|
annexin A4 |
| chr2_+_69727563 | 0.15 |
ENSMUST00000055758.16
ENSMUST00000112251.9 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr10_-_107747995 | 0.15 |
ENSMUST00000165341.5
|
Otogl
|
otogelin-like |
| chr11_+_3933699 | 0.15 |
ENSMUST00000063004.14
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr2_+_120807498 | 0.14 |
ENSMUST00000067582.14
|
Tmem62
|
transmembrane protein 62 |
| chr15_+_31225302 | 0.14 |
ENSMUST00000186425.7
|
Dap
|
death-associated protein |
| chr11_-_70560110 | 0.14 |
ENSMUST00000129434.2
ENSMUST00000018431.13 |
Spag7
|
sperm associated antigen 7 |
| chr15_-_55411560 | 0.14 |
ENSMUST00000165356.2
|
Mrpl13
|
mitochondrial ribosomal protein L13 |
| chr14_-_86986541 | 0.13 |
ENSMUST00000226254.2
|
Diaph3
|
diaphanous related formin 3 |
| chrX_+_111221031 | 0.13 |
ENSMUST00000026599.10
ENSMUST00000113415.2 |
Apool
|
apolipoprotein O-like |
| chr7_-_100232276 | 0.13 |
ENSMUST00000152876.3
ENSMUST00000150042.8 ENSMUST00000132888.9 |
Mrpl48
|
mitochondrial ribosomal protein L48 |
| chr5_-_38649291 | 0.13 |
ENSMUST00000129099.8
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr8_-_68270870 | 0.13 |
ENSMUST00000059374.5
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr9_+_113641615 | 0.11 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr8_+_69661683 | 0.10 |
ENSMUST00000212681.2
ENSMUST00000212312.2 |
Zfp930
|
zinc finger protein 930 |
| chr5_+_38233901 | 0.10 |
ENSMUST00000146864.6
|
Stx18
|
syntaxin 18 |
| chr15_+_31224616 | 0.10 |
ENSMUST00000186547.7
|
Dap
|
death-associated protein |
| chr15_+_31224460 | 0.10 |
ENSMUST00000044524.16
|
Dap
|
death-associated protein |
| chr17_-_79292856 | 0.09 |
ENSMUST00000118991.2
|
Prkd3
|
protein kinase D3 |
| chr15_+_79578141 | 0.09 |
ENSMUST00000230898.2
ENSMUST00000229046.2 |
Gtpbp1
|
GTP binding protein 1 |
| chr3_+_85946145 | 0.09 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr2_+_152907875 | 0.09 |
ENSMUST00000238488.2
ENSMUST00000129377.8 ENSMUST00000109800.2 |
Ccm2l
|
cerebral cavernous malformation 2-like |
| chr18_+_20643325 | 0.08 |
ENSMUST00000070892.8
ENSMUST00000234945.2 |
Dsg3
|
desmoglein 3 |
| chr9_+_111011388 | 0.08 |
ENSMUST00000217117.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr1_-_179572765 | 0.08 |
ENSMUST00000211943.3
ENSMUST00000131716.4 ENSMUST00000221136.2 |
Kif28
|
kinesin family member 28 |
| chr15_+_99291455 | 0.08 |
ENSMUST00000162624.8
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr2_+_152873772 | 0.08 |
ENSMUST00000037235.7
|
Xkr7
|
X-linked Kx blood group related 7 |
| chr18_+_37651393 | 0.08 |
ENSMUST00000097609.3
|
Pcdhb22
|
protocadherin beta 22 |
| chr8_-_68270936 | 0.07 |
ENSMUST00000120071.8
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr7_-_101486983 | 0.07 |
ENSMUST00000185929.2
ENSMUST00000165052.8 |
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr4_+_150938376 | 0.07 |
ENSMUST00000073600.9
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr4_-_42168603 | 0.07 |
ENSMUST00000098121.4
|
Gm13305
|
predicted gene 13305 |
| chr12_+_84463970 | 0.07 |
ENSMUST00000183146.2
|
Rnf113a2
|
ring finger protein 113A2 |
| chr19_-_38031774 | 0.07 |
ENSMUST00000226068.2
|
Myof
|
myoferlin |
| chr7_+_4340708 | 0.07 |
ENSMUST00000006792.6
ENSMUST00000126417.3 |
Ncr1
|
natural cytotoxicity triggering receptor 1 |
| chr2_+_110427643 | 0.06 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chr3_-_137837117 | 0.06 |
ENSMUST00000029805.13
|
Mttp
|
microsomal triglyceride transfer protein |
| chr13_-_76091931 | 0.06 |
ENSMUST00000022078.12
ENSMUST00000109606.3 |
Rhobtb3
|
Rho-related BTB domain containing 3 |
| chr4_-_42665763 | 0.06 |
ENSMUST00000238770.2
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr17_-_35265702 | 0.06 |
ENSMUST00000097338.11
|
Msh5
|
mutS homolog 5 |
| chr2_+_3771709 | 0.06 |
ENSMUST00000177037.2
|
Fam107b
|
family with sequence similarity 107, member B |
| chr5_-_137784943 | 0.06 |
ENSMUST00000132726.2
|
Mepce
|
methylphosphate capping enzyme |
| chr14_+_75368939 | 0.05 |
ENSMUST00000125833.8
ENSMUST00000124499.8 |
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr1_-_168259839 | 0.05 |
ENSMUST00000188912.7
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr3_+_84859453 | 0.05 |
ENSMUST00000029727.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr19_-_38032006 | 0.05 |
ENSMUST00000172095.3
ENSMUST00000041475.16 |
Myof
|
myoferlin |
| chr14_-_30973164 | 0.05 |
ENSMUST00000226565.2
ENSMUST00000022459.5 |
Phf7
|
PHD finger protein 7 |
| chr3_+_108479015 | 0.05 |
ENSMUST00000143054.2
|
Taf13
|
TATA-box binding protein associated factor 13 |
| chr2_-_34951443 | 0.05 |
ENSMUST00000028233.7
|
Hc
|
hemolytic complement |
| chr15_+_99291491 | 0.04 |
ENSMUST00000159531.3
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr18_+_56695515 | 0.04 |
ENSMUST00000130163.8
ENSMUST00000132628.8 |
Phax
|
phosphorylated adaptor for RNA export |
| chr9_+_53678801 | 0.04 |
ENSMUST00000048670.10
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr18_+_11766333 | 0.04 |
ENSMUST00000115861.9
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chr1_-_4430481 | 0.04 |
ENSMUST00000027032.6
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr17_+_79919267 | 0.04 |
ENSMUST00000223924.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr19_+_29902506 | 0.04 |
ENSMUST00000120388.9
ENSMUST00000144528.8 ENSMUST00000177518.8 |
Il33
|
interleukin 33 |
| chr4_-_32923505 | 0.04 |
ENSMUST00000084749.8
|
Ankrd6
|
ankyrin repeat domain 6 |
| chr11_+_114566257 | 0.04 |
ENSMUST00000045779.6
|
Ttyh2
|
tweety family member 2 |
| chr4_-_32923389 | 0.04 |
ENSMUST00000035719.11
|
Ankrd6
|
ankyrin repeat domain 6 |
| chr19_-_33764859 | 0.03 |
ENSMUST00000148137.9
|
Lipo1
|
lipase, member O1 |
| chr3_-_103553347 | 0.03 |
ENSMUST00000117271.3
|
Atg4a-ps
|
autophagy related 4A, pseudogene |
| chr12_+_86999366 | 0.03 |
ENSMUST00000191463.2
|
Cipc
|
CLOCK interacting protein, circadian |
| chr4_+_109092459 | 0.03 |
ENSMUST00000106631.9
|
Calr4
|
calreticulin 4 |
| chr7_+_30193047 | 0.03 |
ENSMUST00000058280.13
ENSMUST00000133318.8 ENSMUST00000142575.8 ENSMUST00000131040.2 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chrX_-_142716085 | 0.03 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr15_+_31224555 | 0.03 |
ENSMUST00000186109.2
|
Dap
|
death-associated protein |
| chr14_-_75368321 | 0.03 |
ENSMUST00000022574.5
|
Lrrc63
|
leucine rich repeat containing 63 |
| chr8_+_94537910 | 0.03 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr7_+_91321500 | 0.03 |
ENSMUST00000238619.2
ENSMUST00000238467.2 |
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr6_-_141719536 | 0.03 |
ENSMUST00000148411.2
|
Gm5724
|
predicted gene 5724 |
| chr3_-_72875187 | 0.03 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr2_+_85868891 | 0.03 |
ENSMUST00000218397.2
|
Olfr1033
|
olfactory receptor 1033 |
| chr13_+_24023428 | 0.03 |
ENSMUST00000091698.12
ENSMUST00000110422.3 ENSMUST00000166467.9 |
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr12_-_8589545 | 0.03 |
ENSMUST00000095863.10
ENSMUST00000165657.3 |
Slc7a15
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 |
| chr12_-_64521464 | 0.03 |
ENSMUST00000059833.8
|
Fscb
|
fibrous sheath CABYR binding protein |
| chr17_+_12597490 | 0.03 |
ENSMUST00000014578.7
|
Plg
|
plasminogen |
| chr2_+_86122799 | 0.03 |
ENSMUST00000217166.2
|
Olfr1052
|
olfactory receptor 1052 |
| chr2_-_114005752 | 0.02 |
ENSMUST00000102543.5
ENSMUST00000043160.13 |
Aqr
|
aquarius |
| chr9_+_55448432 | 0.02 |
ENSMUST00000034869.11
|
Isl2
|
insulin related protein 2 (islet 2) |
| chr15_-_96929086 | 0.02 |
ENSMUST00000230086.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr1_-_138102972 | 0.02 |
ENSMUST00000195533.6
ENSMUST00000183301.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr2_+_89642395 | 0.02 |
ENSMUST00000214508.2
|
Olfr1255
|
olfactory receptor 1255 |
| chr2_+_69869459 | 0.02 |
ENSMUST00000060208.11
|
Myo3b
|
myosin IIIB |
| chr5_-_74863514 | 0.02 |
ENSMUST00000117388.8
|
Lnx1
|
ligand of numb-protein X 1 |
| chr2_-_63014514 | 0.02 |
ENSMUST00000112452.2
|
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr2_-_86276348 | 0.02 |
ENSMUST00000216165.2
|
Olfr1065
|
olfactory receptor 1065 |
| chr9_+_7571397 | 0.02 |
ENSMUST00000120900.8
ENSMUST00000151853.8 ENSMUST00000152878.3 |
Mmp27
|
matrix metallopeptidase 27 |
| chr15_-_60793115 | 0.02 |
ENSMUST00000096418.5
|
A1bg
|
alpha-1-B glycoprotein |
| chr7_-_108480811 | 0.02 |
ENSMUST00000059617.2
|
Olfr518
|
olfactory receptor 518 |
| chr2_-_140513382 | 0.02 |
ENSMUST00000110057.3
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr5_+_35198853 | 0.02 |
ENSMUST00000030985.10
ENSMUST00000202573.2 |
Hgfac
|
hepatocyte growth factor activator |
| chr1_-_126758369 | 0.02 |
ENSMUST00000112583.8
ENSMUST00000094609.10 |
Nckap5
|
NCK-associated protein 5 |
| chrX_+_162923474 | 0.02 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr4_+_101365144 | 0.02 |
ENSMUST00000149047.8
ENSMUST00000106929.10 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr4_+_138694422 | 0.02 |
ENSMUST00000116094.5
ENSMUST00000239443.2 |
Rnf186
|
ring finger protein 186 |
| chr1_-_126758520 | 0.02 |
ENSMUST00000162646.8
|
Nckap5
|
NCK-associated protein 5 |
| chr16_+_16688692 | 0.02 |
ENSMUST00000232547.2
|
Top3b
|
topoisomerase (DNA) III beta |
| chr12_+_59142439 | 0.02 |
ENSMUST00000219140.3
|
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr19_+_13316226 | 0.02 |
ENSMUST00000207124.3
|
Olfr1466
|
olfactory receptor 1466 |
| chr4_+_109092610 | 0.02 |
ENSMUST00000106628.8
|
Calr4
|
calreticulin 4 |
| chr1_+_74700952 | 0.02 |
ENSMUST00000129890.8
|
Ttll4
|
tubulin tyrosine ligase-like family, member 4 |
| chr2_-_134396268 | 0.02 |
ENSMUST00000028704.3
|
Hao1
|
hydroxyacid oxidase 1, liver |
| chr7_-_28661751 | 0.02 |
ENSMUST00000068045.14
ENSMUST00000217157.2 |
Actn4
|
actinin alpha 4 |
| chr2_+_67004178 | 0.02 |
ENSMUST00000239009.2
ENSMUST00000238912.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr3_-_33136153 | 0.02 |
ENSMUST00000108225.10
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr2_+_109522781 | 0.02 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr7_-_23536797 | 0.02 |
ENSMUST00000226319.2
ENSMUST00000228793.2 ENSMUST00000228280.2 ENSMUST00000227661.2 ENSMUST00000226767.2 ENSMUST00000227129.2 |
Vmn1r176
|
vomeronasal 1 receptor 176 |
| chr10_-_129593612 | 0.02 |
ENSMUST00000213379.2
ENSMUST00000217106.2 |
Olfr807
|
olfactory receptor 807 |
| chr4_-_88798438 | 0.02 |
ENSMUST00000056014.3
|
Ifne
|
interferon epsilon |
| chr6_+_65502344 | 0.02 |
ENSMUST00000212375.2
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chr14_+_75368532 | 0.02 |
ENSMUST00000143539.8
ENSMUST00000134114.8 |
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr2_-_87467879 | 0.02 |
ENSMUST00000216082.2
|
Olfr1132
|
olfactory receptor 1132 |
| chr14_+_96118660 | 0.02 |
ENSMUST00000228913.2
ENSMUST00000045892.3 |
Spertl
|
spermatid associated like |
| chr4_+_115741581 | 0.02 |
ENSMUST00000097918.3
|
Kncn
|
kinocilin |
| chr1_-_158642039 | 0.02 |
ENSMUST00000161589.3
|
Pappa2
|
pappalysin 2 |
| chr18_+_32948436 | 0.01 |
ENSMUST00000025237.5
|
Tslp
|
thymic stromal lymphopoietin |
| chr6_+_142244145 | 0.01 |
ENSMUST00000041993.3
|
Iapp
|
islet amyloid polypeptide |
| chr13_+_24023386 | 0.01 |
ENSMUST00000039721.14
|
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr7_-_107759078 | 0.01 |
ENSMUST00000208296.2
|
Olfr485
|
olfactory receptor 485 |
| chr1_-_135934080 | 0.01 |
ENSMUST00000166193.9
|
Igfn1
|
immunoglobulin-like and fibronectin type III domain containing 1 |
| chr10_+_129357661 | 0.01 |
ENSMUST00000217228.2
|
Olfr791
|
olfactory receptor 791 |
| chr7_+_102420428 | 0.01 |
ENSMUST00000213432.2
|
Olfr561
|
olfactory receptor 561 |
| chr2_+_111610658 | 0.01 |
ENSMUST00000054004.2
|
Olfr1302
|
olfactory receptor 1302 |
| chr6_-_30936013 | 0.01 |
ENSMUST00000101589.5
|
Klf14
|
Kruppel-like factor 14 |
| chr15_-_98507913 | 0.01 |
ENSMUST00000226500.2
ENSMUST00000227501.2 |
Adcy6
|
adenylate cyclase 6 |
| chr2_+_85648823 | 0.01 |
ENSMUST00000214416.2
|
Olfr1018
|
olfactory receptor 1018 |
| chr5_+_14075281 | 0.01 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr4_+_151004636 | 0.01 |
ENSMUST00000030808.10
|
Tnfrsf9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr13_+_21316868 | 0.01 |
ENSMUST00000096006.3
|
Olfr263
|
olfactory receptor 263 |
| chr1_-_134477400 | 0.01 |
ENSMUST00000172898.2
|
Mgat4e
|
MGAT4 family, member E |
| chr2_+_162829250 | 0.01 |
ENSMUST00000018012.14
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr9_+_38435026 | 0.01 |
ENSMUST00000074987.3
|
Olfr911-ps1
|
olfactory receptor 911, pseudogene 1 |
| chr10_-_129591354 | 0.01 |
ENSMUST00000059038.3
|
Olfr807
|
olfactory receptor 807 |
| chr5_-_87638728 | 0.01 |
ENSMUST00000147854.6
|
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr3_-_69767208 | 0.01 |
ENSMUST00000171529.4
ENSMUST00000051239.13 |
Sptssb
|
serine palmitoyltransferase, small subunit B |
| chr5_-_28672091 | 0.01 |
ENSMUST00000002708.5
|
Shh
|
sonic hedgehog |
| chr1_-_138103021 | 0.01 |
ENSMUST00000182755.8
ENSMUST00000193650.2 ENSMUST00000182283.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr17_-_31348576 | 0.01 |
ENSMUST00000024827.5
|
Tff3
|
trefoil factor 3, intestinal |
| chr17_-_43813664 | 0.01 |
ENSMUST00000024707.9
ENSMUST00000117137.8 |
Mep1a
|
meprin 1 alpha |
| chr3_+_75982890 | 0.01 |
ENSMUST00000160261.8
|
Fstl5
|
follistatin-like 5 |
| chr3_-_98364359 | 0.01 |
ENSMUST00000188356.3
ENSMUST00000167753.8 |
Gm4450
|
predicted gene 4450 |
| chr17_+_35960600 | 0.01 |
ENSMUST00000171166.3
|
Sfta2
|
surfactant associated 2 |
| chr1_-_172722589 | 0.01 |
ENSMUST00000027824.7
|
Apcs
|
serum amyloid P-component |
| chr2_-_140513320 | 0.01 |
ENSMUST00000056760.4
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr3_-_144738526 | 0.01 |
ENSMUST00000029919.7
|
Clca1
|
chloride channel accessory 1 |
| chr16_-_88671157 | 0.01 |
ENSMUST00000076186.4
|
Krtap19-2
|
keratin associated protein 19-2 |
| chr3_+_14545751 | 0.01 |
ENSMUST00000037321.8
ENSMUST00000120484.8 ENSMUST00000120801.2 |
Slc7a12
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr19_+_58500411 | 0.01 |
ENSMUST00000235305.2
ENSMUST00000069419.8 |
Ccdc172
|
coiled-coil domain containing 172 |
| chr2_+_81883566 | 0.01 |
ENSMUST00000047527.8
|
Zfp804a
|
zinc finger protein 804A |
| chr13_-_24098951 | 0.01 |
ENSMUST00000021769.16
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr7_-_104856137 | 0.01 |
ENSMUST00000214147.2
|
Olfr686
|
olfactory receptor 686 |
| chr13_-_24098981 | 0.01 |
ENSMUST00000110407.4
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr2_-_86218905 | 0.01 |
ENSMUST00000213998.2
|
Olfr1058
|
olfactory receptor 1058 |
| chr3_+_90444537 | 0.01 |
ENSMUST00000098911.10
|
S100a16
|
S100 calcium binding protein A16 |
| chr3_+_90444613 | 0.01 |
ENSMUST00000107335.2
|
S100a16
|
S100 calcium binding protein A16 |
| chr19_+_30210320 | 0.01 |
ENSMUST00000025797.7
|
Mbl2
|
mannose-binding lectin (protein C) 2 |
| chr12_+_87541491 | 0.01 |
ENSMUST00000220499.2
|
Eif1ad12
|
eukaryotic translation initiation factor 1A domain containing 12 |
| chr6_+_21986445 | 0.01 |
ENSMUST00000115382.8
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr2_+_87405430 | 0.01 |
ENSMUST00000081034.2
|
Olfr1129
|
olfactory receptor 1129 |
| chr10_+_29074950 | 0.01 |
ENSMUST00000217011.2
|
Gm49353
|
predicted gene, 49353 |
| chr14_+_50690511 | 0.01 |
ENSMUST00000089838.2
|
Olfr740
|
olfactory receptor 740 |
| chr9_-_70842090 | 0.01 |
ENSMUST00000034731.10
|
Lipc
|
lipase, hepatic |
| chr4_+_151004606 | 0.01 |
ENSMUST00000105672.10
|
Tnfrsf9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr14_+_33073249 | 0.01 |
ENSMUST00000140711.3
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr10_-_88440869 | 0.01 |
ENSMUST00000119185.8
ENSMUST00000238199.2 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr12_+_29584560 | 0.01 |
ENSMUST00000021009.10
|
Myt1l
|
myelin transcription factor 1-like |
| chr3_-_144514386 | 0.01 |
ENSMUST00000197013.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr5_+_7354130 | 0.01 |
ENSMUST00000160634.2
ENSMUST00000159546.2 |
Tex47
|
testis expressed 47 |
| chr1_+_164103127 | 0.01 |
ENSMUST00000027867.7
|
Ccdc181
|
coiled-coil domain containing 181 |
| chr4_+_109092829 | 0.01 |
ENSMUST00000030285.8
|
Calr4
|
calreticulin 4 |
| chr11_-_41891111 | 0.01 |
ENSMUST00000109290.2
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr10_-_129811436 | 0.01 |
ENSMUST00000170920.2
|
Olfr247
|
olfactory receptor 247 |
| chr3_+_153549846 | 0.01 |
ENSMUST00000044089.4
|
Asb17
|
ankyrin repeat and SOCS box-containing 17 |
| chr5_-_66672158 | 0.01 |
ENSMUST00000161879.8
ENSMUST00000159357.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr2_+_87663385 | 0.01 |
ENSMUST00000077580.2
|
Olfr1148
|
olfactory receptor 1148 |
| chr10_+_90412114 | 0.01 |
ENSMUST00000182427.8
ENSMUST00000182053.8 ENSMUST00000182113.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr6_+_42733338 | 0.01 |
ENSMUST00000215796.2
|
Olfr38
|
olfactory receptor 38 |
| chr19_+_13700642 | 0.01 |
ENSMUST00000207208.3
ENSMUST00000208667.3 |
Olfr1493-ps1
|
olfactory receptor 1493, pseudogene 1 |
| chr10_-_38697775 | 0.01 |
ENSMUST00000179279.2
|
Rfpl4b
|
ret finger protein-like 4B |
| chr3_-_144638284 | 0.01 |
ENSMUST00000098549.4
|
Clca4b
|
chloride channel accessory 4B |
| chr6_-_38101503 | 0.01 |
ENSMUST00000040259.8
|
Atp6v0a4
|
ATPase, H+ transporting, lysosomal V0 subunit A4 |
| chr9_-_35429565 | 0.01 |
ENSMUST00000183573.5
|
Gm1113
|
predicted gene 1113 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfhx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.1 | 0.4 | GO:0015886 | heme transport(GO:0015886) methotrexate transport(GO:0051958) |
| 0.0 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.7 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.4 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.3 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0031437 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.8 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.5 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.0 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.0 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.0 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.1 | GO:1904511 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 0.9 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.1 | 0.4 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.1 | 0.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.5 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |