Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
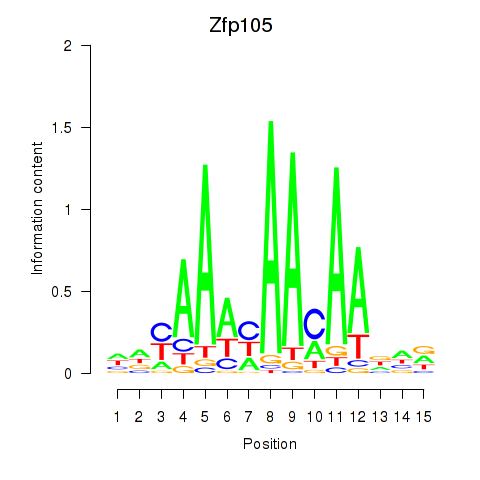
Results for Zfp105
Z-value: 0.32
Transcription factors associated with Zfp105
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp105
|
ENSMUSG00000057895.12 | zinc finger protein 105 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp105 | mm39_v1_chr9_+_122752164_122752176 | 0.14 | 8.3e-01 | Click! |
Activity profile of Zfp105 motif
Sorted Z-values of Zfp105 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_45890303 | 0.40 |
ENSMUST00000178561.8
|
Ccdc180
|
coiled-coil domain containing 180 |
| chr5_+_146769700 | 0.23 |
ENSMUST00000035983.12
|
Rpl21
|
ribosomal protein L21 |
| chrM_+_3906 | 0.22 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr15_-_50753437 | 0.21 |
ENSMUST00000077935.6
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr6_-_99412306 | 0.16 |
ENSMUST00000113322.9
ENSMUST00000176850.8 ENSMUST00000176632.8 |
Foxp1
|
forkhead box P1 |
| chr7_-_15781838 | 0.16 |
ENSMUST00000210781.2
|
Bicra
|
BRD4 interacting chromatin remodeling complex associated protein |
| chr15_-_50753061 | 0.15 |
ENSMUST00000165201.9
ENSMUST00000184458.8 |
Trps1
|
transcriptional repressor GATA binding 1 |
| chr15_-_50752437 | 0.13 |
ENSMUST00000183997.8
ENSMUST00000183757.8 |
Trps1
|
transcriptional repressor GATA binding 1 |
| chr6_-_99005835 | 0.13 |
ENSMUST00000154163.9
|
Foxp1
|
forkhead box P1 |
| chr11_+_108811168 | 0.12 |
ENSMUST00000052915.14
|
Axin2
|
axin 2 |
| chr6_-_142910094 | 0.09 |
ENSMUST00000032421.4
|
St8sia1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr6_+_135339543 | 0.09 |
ENSMUST00000205156.3
|
Emp1
|
epithelial membrane protein 1 |
| chr5_-_143133260 | 0.09 |
ENSMUST00000215102.2
ENSMUST00000213631.2 ENSMUST00000164536.5 |
Olfr718-ps1
|
olfactory receptor 718, pseudogene 1 |
| chr6_+_135339929 | 0.08 |
ENSMUST00000032330.16
|
Emp1
|
epithelial membrane protein 1 |
| chr11_+_44508137 | 0.08 |
ENSMUST00000109268.2
ENSMUST00000101326.10 ENSMUST00000081265.12 |
Ebf1
|
early B cell factor 1 |
| chr10_-_111833138 | 0.07 |
ENSMUST00000074805.12
|
Glipr1
|
GLI pathogenesis-related 1 (glioma) |
| chr9_+_21867043 | 0.07 |
ENSMUST00000053583.7
|
Swsap1
|
SWIM type zinc finger 7 associated protein 1 |
| chr3_+_20043315 | 0.06 |
ENSMUST00000173779.2
|
Cp
|
ceruloplasmin |
| chr3_+_138233004 | 0.06 |
ENSMUST00000196990.5
ENSMUST00000200239.5 ENSMUST00000200100.2 |
Eif4e
|
eukaryotic translation initiation factor 4E |
| chr7_-_102638531 | 0.05 |
ENSMUST00000215606.2
|
Olfr578
|
olfactory receptor 578 |
| chr10_-_30647881 | 0.05 |
ENSMUST00000215740.2
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr15_-_78004211 | 0.04 |
ENSMUST00000019290.3
|
Cacng2
|
calcium channel, voltage-dependent, gamma subunit 2 |
| chr2_-_87985537 | 0.04 |
ENSMUST00000216951.2
|
Olfr1167
|
olfactory receptor 1167 |
| chr6_-_53797748 | 0.04 |
ENSMUST00000127748.5
|
Tril
|
TLR4 interactor with leucine-rich repeats |
| chr3_-_116388334 | 0.03 |
ENSMUST00000197190.5
ENSMUST00000198454.2 |
Trmt13
|
tRNA methyltransferase 13 |
| chr10_-_30647836 | 0.03 |
ENSMUST00000215926.2
ENSMUST00000213836.2 |
Ncoa7
|
nuclear receptor coactivator 7 |
| chr12_-_52018072 | 0.03 |
ENSMUST00000040583.7
|
Heatr5a
|
HEAT repeat containing 5A |
| chr6_+_21949986 | 0.02 |
ENSMUST00000149728.7
|
Ing3
|
inhibitor of growth family, member 3 |
| chr2_+_143388062 | 0.02 |
ENSMUST00000028905.10
|
Pcsk2
|
proprotein convertase subtilisin/kexin type 2 |
| chr7_-_34012934 | 0.02 |
ENSMUST00000206399.2
|
Garre1
|
granule associated Rac and RHOG effector 1 |
| chr2_-_65397850 | 0.02 |
ENSMUST00000238483.2
ENSMUST00000100069.9 |
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr4_+_134042423 | 0.02 |
ENSMUST00000105875.8
ENSMUST00000030638.7 |
Trim63
|
tripartite motif-containing 63 |
| chr10_-_78427721 | 0.02 |
ENSMUST00000040580.7
|
Syde1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
| chr2_+_107120934 | 0.01 |
ENSMUST00000037012.3
|
Kcna4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr12_+_87921198 | 0.01 |
ENSMUST00000110145.12
ENSMUST00000181843.2 ENSMUST00000180706.8 ENSMUST00000181394.8 ENSMUST00000181326.8 ENSMUST00000181300.2 |
Gm2042
|
predicted gene 2042 |
| chr12_-_84031622 | 0.01 |
ENSMUST00000164935.3
|
Heatr4
|
HEAT repeat containing 4 |
| chr5_+_20112500 | 0.01 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr6_+_97968737 | 0.01 |
ENSMUST00000043628.13
ENSMUST00000203938.2 |
Mitf
|
melanogenesis associated transcription factor |
| chr2_-_151528259 | 0.01 |
ENSMUST00000149319.2
|
Tmem74bos
|
transmembrane 74B, opposite strand |
| chr5_-_23889591 | 0.01 |
ENSMUST00000198549.2
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr14_-_101846551 | 0.01 |
ENSMUST00000100340.4
|
Tbc1d4
|
TBC1 domain family, member 4 |
| chr11_-_99742434 | 0.00 |
ENSMUST00000107437.2
|
Krtap4-16
|
keratin associated protein 4-16 |
| chr11_+_108811626 | 0.00 |
ENSMUST00000140821.2
|
Axin2
|
axin 2 |
| chr2_+_67004178 | 0.00 |
ENSMUST00000239009.2
ENSMUST00000238912.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr8_+_94537910 | 0.00 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr13_-_21823691 | 0.00 |
ENSMUST00000043081.3
|
Olfr11
|
olfactory receptor 11 |
| chr18_-_25886750 | 0.00 |
ENSMUST00000224553.2
ENSMUST00000025117.14 |
Celf4
|
CUGBP, Elav-like family member 4 |
| chr16_-_55103598 | 0.00 |
ENSMUST00000036412.4
|
Zpld1
|
zona pellucida like domain containing 1 |
| chr9_-_117080869 | 0.00 |
ENSMUST00000172564.3
|
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr3_+_63203516 | 0.00 |
ENSMUST00000029400.7
|
Mme
|
membrane metallo endopeptidase |
| chr1_+_120530134 | 0.00 |
ENSMUST00000079721.9
|
En1
|
engrailed 1 |
| chr5_+_17779273 | 0.00 |
ENSMUST00000030568.14
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr3_+_63203235 | 0.00 |
ENSMUST00000194134.6
|
Mme
|
membrane metallo endopeptidase |
| chr10_+_90412114 | 0.00 |
ENSMUST00000182427.8
ENSMUST00000182053.8 ENSMUST00000182113.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr6_+_52290415 | 0.00 |
ENSMUST00000031787.8
ENSMUST00000129243.3 |
Evx1
|
even-skipped homeobox 1 |
| chr7_+_104583447 | 0.00 |
ENSMUST00000214260.2
ENSMUST00000210138.3 |
Olfr669
|
olfactory receptor 669 |
| chr6_+_131520725 | 0.00 |
ENSMUST00000161385.5
|
5430401F13Rik
|
RIKEN cDNA 5430401F13 gene |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp105
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0061181 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.4 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |