Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
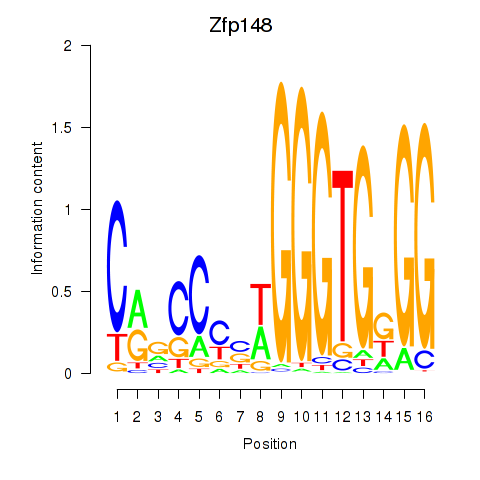
Results for Zfp148
Z-value: 1.15
Transcription factors associated with Zfp148
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp148
|
ENSMUSG00000022811.18 | zinc finger protein 148 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp148 | mm39_v1_chr16_+_33201227_33201275 | 0.99 | 4.6e-04 | Click! |
Activity profile of Zfp148 motif
Sorted Z-values of Zfp148 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_72935382 | 0.89 |
ENSMUST00000144337.2
|
Tmsb10
|
thymosin, beta 10 |
| chr12_-_113271532 | 0.76 |
ENSMUST00000192188.3
ENSMUST00000103418.3 |
Ighg2b
|
immunoglobulin heavy constant gamma 2B |
| chr1_-_162687488 | 0.75 |
ENSMUST00000134098.8
ENSMUST00000111518.3 |
Fmo1
|
flavin containing monooxygenase 1 |
| chr9_-_109678685 | 0.65 |
ENSMUST00000112022.5
|
Camp
|
cathelicidin antimicrobial peptide |
| chr4_-_118795809 | 0.62 |
ENSMUST00000215312.3
|
Olfr1328
|
olfactory receptor 1328 |
| chrX_-_7242065 | 0.54 |
ENSMUST00000191497.2
ENSMUST00000115744.2 |
Usp27x
|
ubiquitin specific peptidase 27, X chromosome |
| chr5_-_116427003 | 0.51 |
ENSMUST00000086483.4
ENSMUST00000050178.13 |
Ccdc60
|
coiled-coil domain containing 60 |
| chr7_+_28834350 | 0.49 |
ENSMUST00000159975.8
ENSMUST00000094617.11 ENSMUST00000032811.12 |
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr9_+_110848339 | 0.48 |
ENSMUST00000198884.5
ENSMUST00000196777.5 ENSMUST00000196209.5 ENSMUST00000035077.8 ENSMUST00000196122.3 |
Ltf
|
lactotransferrin |
| chr11_+_77873535 | 0.44 |
ENSMUST00000108360.8
ENSMUST00000049167.14 |
Phf12
|
PHD finger protein 12 |
| chr7_+_28834276 | 0.43 |
ENSMUST00000161522.8
ENSMUST00000204845.3 ENSMUST00000205027.3 ENSMUST00000204194.3 ENSMUST00000203070.3 ENSMUST00000203380.3 |
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr7_+_96730915 | 0.43 |
ENSMUST00000206791.2
|
Gab2
|
growth factor receptor bound protein 2-associated protein 2 |
| chr10_-_7831657 | 0.43 |
ENSMUST00000147938.2
|
Tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr7_+_3337591 | 0.41 |
ENSMUST00000203328.4
|
Myadm
|
myeloid-associated differentiation marker |
| chr18_-_6135888 | 0.40 |
ENSMUST00000182383.8
ENSMUST00000062584.14 ENSMUST00000077128.13 ENSMUST00000182038.2 ENSMUST00000182213.8 ENSMUST00000182559.8 |
Arhgap12
|
Rho GTPase activating protein 12 |
| chr6_-_41681273 | 0.40 |
ENSMUST00000031899.14
|
Kel
|
Kell blood group |
| chr19_+_5348329 | 0.39 |
ENSMUST00000061169.7
|
Gal3st3
|
galactose-3-O-sulfotransferase 3 |
| chr8_+_91635192 | 0.39 |
ENSMUST00000211403.2
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr7_+_28834391 | 0.38 |
ENSMUST00000160194.8
|
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr15_-_82783978 | 0.38 |
ENSMUST00000230403.2
|
Tcf20
|
transcription factor 20 |
| chr11_+_70453666 | 0.37 |
ENSMUST00000072237.13
ENSMUST00000072873.14 |
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr19_+_60878802 | 0.37 |
ENSMUST00000236876.2
|
Grk5
|
G protein-coupled receptor kinase 5 |
| chr2_-_166904625 | 0.36 |
ENSMUST00000128676.2
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr11_+_60244132 | 0.36 |
ENSMUST00000070805.13
ENSMUST00000094140.9 ENSMUST00000108723.9 ENSMUST00000108722.5 |
Drc3
|
dynein regulatory complex subunit 3 |
| chr10_+_127595590 | 0.36 |
ENSMUST00000073639.6
|
Rdh1
|
retinol dehydrogenase 1 (all trans) |
| chr15_-_75438457 | 0.35 |
ENSMUST00000163116.8
ENSMUST00000023241.12 |
Ly6h
|
lymphocyte antigen 6 complex, locus H |
| chr9_-_97915036 | 0.35 |
ENSMUST00000162295.2
|
Clstn2
|
calsyntenin 2 |
| chr4_-_108637700 | 0.35 |
ENSMUST00000106658.8
|
Zfyve9
|
zinc finger, FYVE domain containing 9 |
| chr6_-_72935468 | 0.34 |
ENSMUST00000114050.8
|
Tmsb10
|
thymosin, beta 10 |
| chr6_-_72935171 | 0.34 |
ENSMUST00000114049.2
|
Tmsb10
|
thymosin, beta 10 |
| chr12_+_105302853 | 0.34 |
ENSMUST00000180458.9
|
Tunar
|
Tcl1 upstream neural differentiation associated RNA |
| chr5_-_129747129 | 0.32 |
ENSMUST00000049778.7
|
Zfp11
|
zinc finger protein 11 |
| chr1_+_166828982 | 0.30 |
ENSMUST00000165874.8
ENSMUST00000190081.7 |
Fam78b
|
family with sequence similarity 78, member B |
| chr4_+_43669266 | 0.29 |
ENSMUST00000107864.8
|
Tmem8b
|
transmembrane protein 8B |
| chr3_+_96604390 | 0.29 |
ENSMUST00000162778.3
ENSMUST00000064900.16 |
Pias3
|
protein inhibitor of activated STAT 3 |
| chr6_-_116170389 | 0.28 |
ENSMUST00000088896.10
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr15_+_84565174 | 0.28 |
ENSMUST00000065499.5
|
Prr5
|
proline rich 5 (renal) |
| chr11_-_60243695 | 0.27 |
ENSMUST00000095254.12
ENSMUST00000102683.11 ENSMUST00000093048.13 ENSMUST00000093046.13 ENSMUST00000064019.15 ENSMUST00000102682.5 |
Tom1l2
|
target of myb1-like 2 (chicken) |
| chr16_+_75389732 | 0.27 |
ENSMUST00000046378.14
ENSMUST00000114249.8 ENSMUST00000114253.2 |
Rbm11
|
RNA binding motif protein 11 |
| chr4_+_118745806 | 0.27 |
ENSMUST00000106361.3
ENSMUST00000105035.3 |
Olfr1330
|
olfactory receptor 1330 |
| chr14_-_25769457 | 0.26 |
ENSMUST00000069180.8
|
Zcchc24
|
zinc finger, CCHC domain containing 24 |
| chr9_+_109760856 | 0.26 |
ENSMUST00000169851.8
|
Map4
|
microtubule-associated protein 4 |
| chr7_+_3339059 | 0.26 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr10_+_127637015 | 0.25 |
ENSMUST00000071646.2
|
Rdh16
|
retinol dehydrogenase 16 |
| chr11_-_69692542 | 0.25 |
ENSMUST00000011285.11
ENSMUST00000102585.2 |
Fgf11
|
fibroblast growth factor 11 |
| chr7_-_132415257 | 0.25 |
ENSMUST00000097999.9
|
Fam53b
|
family with sequence similarity 53, member B |
| chr15_-_68130636 | 0.25 |
ENSMUST00000162173.8
ENSMUST00000160248.8 ENSMUST00000162054.9 |
Zfat
|
zinc finger and AT hook domain containing |
| chr7_+_3648264 | 0.25 |
ENSMUST00000206287.2
ENSMUST00000038913.16 |
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr9_+_110074574 | 0.25 |
ENSMUST00000197850.5
|
Cspg5
|
chondroitin sulfate proteoglycan 5 |
| chr7_+_4467730 | 0.24 |
ENSMUST00000086372.8
ENSMUST00000169820.8 ENSMUST00000163893.8 ENSMUST00000170635.2 |
Eps8l1
|
EPS8-like 1 |
| chr9_-_107749594 | 0.24 |
ENSMUST00000183035.2
|
Rbm6
|
RNA binding motif protein 6 |
| chr4_-_117740624 | 0.24 |
ENSMUST00000030266.12
|
B4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr4_+_62204678 | 0.24 |
ENSMUST00000084530.9
|
Slc31a2
|
solute carrier family 31, member 2 |
| chr5_-_137312424 | 0.24 |
ENSMUST00000199121.2
|
Trip6
|
thyroid hormone receptor interactor 6 |
| chr1_-_133589020 | 0.23 |
ENSMUST00000193504.6
ENSMUST00000195067.2 ENSMUST00000191896.6 ENSMUST00000194668.6 ENSMUST00000195424.6 ENSMUST00000179598.4 ENSMUST00000027736.13 |
Zc3h11a
Gm38394
Zc3h11a
|
zinc finger CCCH type containing 11A predicted gene, 38394 zinc finger CCCH type containing 11A |
| chr2_-_168432235 | 0.23 |
ENSMUST00000109184.8
|
Nfatc2
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 |
| chr7_-_63588610 | 0.23 |
ENSMUST00000063694.10
|
Klf13
|
Kruppel-like factor 13 |
| chr7_+_79675727 | 0.23 |
ENSMUST00000049680.10
|
Zfp710
|
zinc finger protein 710 |
| chr18_-_25887173 | 0.23 |
ENSMUST00000225477.2
|
Celf4
|
CUGBP, Elav-like family member 4 |
| chr12_-_100865783 | 0.22 |
ENSMUST00000053668.10
|
Gpr68
|
G protein-coupled receptor 68 |
| chr12_+_12312135 | 0.22 |
ENSMUST00000069066.14
|
Cyria
|
CYFIP related Rac1 interactor A |
| chr12_+_3856510 | 0.22 |
ENSMUST00000172719.8
|
Dnmt3a
|
DNA methyltransferase 3A |
| chr5_+_30745447 | 0.22 |
ENSMUST00000066295.5
|
Kcnk3
|
potassium channel, subfamily K, member 3 |
| chr9_-_107512511 | 0.22 |
ENSMUST00000192615.6
ENSMUST00000192837.2 ENSMUST00000193876.2 |
Gnai2
|
guanine nucleotide binding protein (G protein), alpha inhibiting 2 |
| chr12_-_35584968 | 0.21 |
ENSMUST00000116436.9
|
Ahr
|
aryl-hydrocarbon receptor |
| chr12_+_108145802 | 0.21 |
ENSMUST00000221167.2
|
Ccnk
|
cyclin K |
| chr11_+_120612278 | 0.21 |
ENSMUST00000018156.12
|
Rac3
|
Rac family small GTPase 3 |
| chrX_+_55391749 | 0.21 |
ENSMUST00000101560.4
|
Zfp449
|
zinc finger protein 449 |
| chr7_+_46045862 | 0.21 |
ENSMUST00000025202.8
|
Kcnc1
|
potassium voltage gated channel, Shaw-related subfamily, member 1 |
| chr8_+_85449632 | 0.21 |
ENSMUST00000098571.5
|
G430095P16Rik
|
RIKEN cDNA G430095P16 gene |
| chr16_+_58490625 | 0.21 |
ENSMUST00000060077.7
|
Cpox
|
coproporphyrinogen oxidase |
| chr12_-_111344139 | 0.21 |
ENSMUST00000041965.5
|
Cdc42bpb
|
CDC42 binding protein kinase beta |
| chr7_-_43139390 | 0.21 |
ENSMUST00000107974.3
|
Iglon5
|
IgLON family member 5 |
| chr15_+_78993111 | 0.21 |
ENSMUST00000040320.10
|
Micall1
|
microtubule associated monooxygenase, calponin and LIM domain containing -like 1 |
| chr11_-_101117237 | 0.20 |
ENSMUST00000100417.3
ENSMUST00000107285.8 ENSMUST00000107284.8 |
Ezh1
|
enhancer of zeste 1 polycomb repressive complex 2 subunit |
| chr2_-_166904902 | 0.20 |
ENSMUST00000048988.14
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr1_+_59802543 | 0.20 |
ENSMUST00000087435.7
|
Bmpr2
|
bone morphogenetic protein receptor, type II (serine/threonine kinase) |
| chr11_-_86248395 | 0.20 |
ENSMUST00000043624.9
|
Med13
|
mediator complex subunit 13 |
| chr7_+_26853139 | 0.20 |
ENSMUST00000164093.8
|
Cyp2t4
|
cytochrome P450, family 2, subfamily t, polypeptide 4 |
| chr18_+_4920513 | 0.20 |
ENSMUST00000126977.8
|
Svil
|
supervillin |
| chr4_-_35225848 | 0.20 |
ENSMUST00000108127.4
|
C9orf72
|
C9orf72, member of C9orf72-SMCR8 complex |
| chr11_-_62348115 | 0.20 |
ENSMUST00000069456.11
ENSMUST00000018645.13 |
Ncor1
|
nuclear receptor co-repressor 1 |
| chr2_+_156681991 | 0.19 |
ENSMUST00000073352.10
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr12_+_12312163 | 0.19 |
ENSMUST00000069005.10
ENSMUST00000223061.2 |
Cyria
|
CYFIP related Rac1 interactor A |
| chr1_+_166829001 | 0.19 |
ENSMUST00000126198.3
|
Fam78b
|
family with sequence similarity 78, member B |
| chr2_-_44817218 | 0.18 |
ENSMUST00000100127.9
|
Gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr8_-_96058383 | 0.18 |
ENSMUST00000057717.8
|
Zfp319
|
zinc finger protein 319 |
| chrX_-_151110425 | 0.18 |
ENSMUST00000195280.3
|
Kantr
|
Kdm5c adjacent non-coding transcript |
| chr2_-_146353911 | 0.18 |
ENSMUST00000109986.9
|
Ralgapa2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr16_+_24212284 | 0.17 |
ENSMUST00000038053.14
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr19_+_45352173 | 0.17 |
ENSMUST00000223764.2
ENSMUST00000065601.13 ENSMUST00000224102.2 ENSMUST00000111936.4 |
Btrc
|
beta-transducin repeat containing protein |
| chr7_+_141276575 | 0.17 |
ENSMUST00000185406.8
|
Muc2
|
mucin 2 |
| chr14_+_47536075 | 0.17 |
ENSMUST00000227554.2
|
Mapk1ip1l
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr7_+_28466160 | 0.17 |
ENSMUST00000122915.8
ENSMUST00000072965.5 ENSMUST00000170068.9 |
Sirt2
|
sirtuin 2 |
| chr3_+_106455114 | 0.17 |
ENSMUST00000067630.13
ENSMUST00000134396.8 ENSMUST00000121034.8 ENSMUST00000029507.13 ENSMUST00000144746.8 ENSMUST00000132923.8 ENSMUST00000151465.2 |
Dram2
|
DNA-damage regulated autophagy modulator 2 |
| chr5_+_34683141 | 0.16 |
ENSMUST00000125817.8
ENSMUST00000067638.14 |
Sh3bp2
|
SH3-domain binding protein 2 |
| chr11_+_101169918 | 0.16 |
ENSMUST00000103107.5
|
Cntd1
|
cyclin N-terminal domain containing 1 |
| chr6_+_103487973 | 0.16 |
ENSMUST00000066905.9
|
Chl1
|
cell adhesion molecule L1-like |
| chr16_+_33504908 | 0.16 |
ENSMUST00000126532.2
|
Heg1
|
heart development protein with EGF-like domains 1 |
| chr4_+_125918333 | 0.16 |
ENSMUST00000106162.8
|
Csf3r
|
colony stimulating factor 3 receptor (granulocyte) |
| chr9_-_107512566 | 0.16 |
ENSMUST00000055704.12
|
Gnai2
|
guanine nucleotide binding protein (G protein), alpha inhibiting 2 |
| chr1_-_132953068 | 0.16 |
ENSMUST00000186617.7
ENSMUST00000067429.10 ENSMUST00000067398.13 ENSMUST00000188090.7 |
Mdm4
|
transformed mouse 3T3 cell double minute 4 |
| chr11_+_75542328 | 0.16 |
ENSMUST00000069057.13
|
Myo1c
|
myosin IC |
| chr11_-_84719779 | 0.16 |
ENSMUST00000047560.8
|
Dhrs11
|
dehydrogenase/reductase (SDR family) member 11 |
| chr2_+_163444214 | 0.16 |
ENSMUST00000171696.8
ENSMUST00000109408.10 |
Ttpal
|
tocopherol (alpha) transfer protein-like |
| chr14_+_121272950 | 0.16 |
ENSMUST00000026635.8
|
Farp1
|
FERM, RhoGEF (Arhgef) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr1_+_118317153 | 0.16 |
ENSMUST00000189738.7
ENSMUST00000187713.7 ENSMUST00000165223.8 ENSMUST00000189570.7 ENSMUST00000191445.7 ENSMUST00000189262.7 |
Clasp1
|
CLIP associating protein 1 |
| chr7_-_30672824 | 0.16 |
ENSMUST00000147431.2
ENSMUST00000098553.11 ENSMUST00000108116.10 |
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr2_+_163444248 | 0.15 |
ENSMUST00000152135.8
|
Ttpal
|
tocopherol (alpha) transfer protein-like |
| chr12_+_108145997 | 0.15 |
ENSMUST00000101055.5
|
Ccnk
|
cyclin K |
| chr9_+_65816206 | 0.15 |
ENSMUST00000205379.2
ENSMUST00000206048.2 ENSMUST00000034949.10 ENSMUST00000154589.2 |
Csnk1g1
|
casein kinase 1, gamma 1 |
| chr13_+_42205491 | 0.15 |
ENSMUST00000060148.6
|
Hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr12_-_4527138 | 0.15 |
ENSMUST00000085814.5
|
Ncoa1
|
nuclear receptor coactivator 1 |
| chr4_-_35225775 | 0.15 |
ENSMUST00000108126.8
|
C9orf72
|
C9orf72, member of C9orf72-SMCR8 complex |
| chr9_+_44394080 | 0.15 |
ENSMUST00000220303.2
|
Bcl9l
|
B cell CLL/lymphoma 9-like |
| chr7_+_16044336 | 0.15 |
ENSMUST00000136781.2
|
Bbc3
|
BCL2 binding component 3 |
| chr9_+_65816370 | 0.15 |
ENSMUST00000206594.2
|
Csnk1g1
|
casein kinase 1, gamma 1 |
| chr10_+_79690452 | 0.15 |
ENSMUST00000165704.8
|
Ptbp1
|
polypyrimidine tract binding protein 1 |
| chr9_-_40366966 | 0.15 |
ENSMUST00000165104.8
ENSMUST00000045682.7 |
Gramd1b
|
GRAM domain containing 1B |
| chr11_+_49094119 | 0.15 |
ENSMUST00000109198.8
ENSMUST00000137061.9 |
Zfp62
|
zinc finger protein 62 |
| chr7_+_92524495 | 0.14 |
ENSMUST00000207594.2
|
Prcp
|
prolylcarboxypeptidase (angiotensinase C) |
| chr17_-_24752683 | 0.14 |
ENSMUST00000061764.14
|
Rab26
|
RAB26, member RAS oncogene family |
| chr16_-_18052937 | 0.14 |
ENSMUST00000076957.7
|
Zdhhc8
|
zinc finger, DHHC domain containing 8 |
| chr11_-_46057224 | 0.14 |
ENSMUST00000020679.3
|
Nipal4
|
NIPA-like domain containing 4 |
| chrX_+_72760183 | 0.14 |
ENSMUST00000002084.14
|
Abcd1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr7_-_46445305 | 0.14 |
ENSMUST00000107653.8
ENSMUST00000107654.8 ENSMUST00000014562.14 ENSMUST00000152759.8 |
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr18_-_25886908 | 0.14 |
ENSMUST00000115816.3
ENSMUST00000223704.2 |
Celf4
|
CUGBP, Elav-like family member 4 |
| chr11_+_104122216 | 0.14 |
ENSMUST00000106992.10
|
Mapt
|
microtubule-associated protein tau |
| chr10_+_126814542 | 0.14 |
ENSMUST00000105256.10
|
Ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr3_+_96604415 | 0.13 |
ENSMUST00000107077.4
|
Pias3
|
protein inhibitor of activated STAT 3 |
| chr8_+_75720286 | 0.13 |
ENSMUST00000211863.2
|
Hmgxb4
|
HMG box domain containing 4 |
| chr14_-_100521888 | 0.13 |
ENSMUST00000226774.2
|
Klf12
|
Kruppel-like factor 12 |
| chr2_+_84670956 | 0.13 |
ENSMUST00000111625.2
|
Slc43a1
|
solute carrier family 43, member 1 |
| chr7_+_92524460 | 0.13 |
ENSMUST00000076052.8
|
Prcp
|
prolylcarboxypeptidase (angiotensinase C) |
| chr19_+_45352514 | 0.13 |
ENSMUST00000224318.2
ENSMUST00000223684.2 |
Btrc
|
beta-transducin repeat containing protein |
| chr10_+_3316505 | 0.13 |
ENSMUST00000217573.2
|
Ppp1r14c
|
protein phosphatase 1, regulatory inhibitor subunit 14C |
| chr7_+_138968988 | 0.13 |
ENSMUST00000106098.8
ENSMUST00000026550.14 |
Inpp5a
|
inositol polyphosphate-5-phosphatase A |
| chr17_-_35340189 | 0.12 |
ENSMUST00000025246.13
ENSMUST00000173114.8 |
Csnk2b
|
casein kinase 2, beta polypeptide |
| chr3_+_90173813 | 0.12 |
ENSMUST00000098914.10
|
Dennd4b
|
DENN/MADD domain containing 4B |
| chr17_-_30795136 | 0.12 |
ENSMUST00000079924.8
ENSMUST00000236584.2 ENSMUST00000236825.2 ENSMUST00000235587.2 |
Btbd9
|
BTB (POZ) domain containing 9 |
| chr4_-_103071988 | 0.12 |
ENSMUST00000036195.13
|
Slc35d1
|
solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 |
| chr2_+_84670543 | 0.12 |
ENSMUST00000111624.8
|
Slc43a1
|
solute carrier family 43, member 1 |
| chr1_+_135075377 | 0.12 |
ENSMUST00000125774.2
|
Arl8a
|
ADP-ribosylation factor-like 8A |
| chr14_+_20745024 | 0.12 |
ENSMUST00000048016.3
|
Fut11
|
fucosyltransferase 11 |
| chr14_-_62998561 | 0.12 |
ENSMUST00000053959.7
ENSMUST00000223585.2 |
Ints6
|
integrator complex subunit 6 |
| chr5_+_128813232 | 0.12 |
ENSMUST00000086056.8
|
Piwil1
|
piwi-like RNA-mediated gene silencing 1 |
| chr7_-_46445085 | 0.12 |
ENSMUST00000123725.2
|
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr7_-_126483851 | 0.12 |
ENSMUST00000071268.11
ENSMUST00000117394.2 |
Taok2
|
TAO kinase 2 |
| chr5_+_110324734 | 0.12 |
ENSMUST00000139611.8
ENSMUST00000031477.9 |
Golga3
|
golgi autoantigen, golgin subfamily a, 3 |
| chr4_+_136038263 | 0.11 |
ENSMUST00000105850.8
ENSMUST00000148843.10 |
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr3_+_104772378 | 0.11 |
ENSMUST00000200132.5
ENSMUST00000123876.6 |
St7l
|
suppression of tumorigenicity 7-like |
| chr7_-_80453033 | 0.11 |
ENSMUST00000167377.3
|
Iqgap1
|
IQ motif containing GTPase activating protein 1 |
| chr4_-_133480922 | 0.11 |
ENSMUST00000145664.9
ENSMUST00000105897.10 |
Arid1a
|
AT rich interactive domain 1A (SWI-like) |
| chr12_+_113115632 | 0.11 |
ENSMUST00000006523.12
ENSMUST00000200553.2 |
Crip1
|
cysteine-rich protein 1 (intestinal) |
| chr17_+_28491085 | 0.11 |
ENSMUST00000169040.3
|
Ppard
|
peroxisome proliferator activator receptor delta |
| chr17_+_35816915 | 0.10 |
ENSMUST00000025271.17
|
Pou5f1
|
POU domain, class 5, transcription factor 1 |
| chr11_+_83193495 | 0.10 |
ENSMUST00000176430.8
ENSMUST00000065692.14 ENSMUST00000142680.2 |
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr10_+_3316057 | 0.10 |
ENSMUST00000043374.7
|
Ppp1r14c
|
protein phosphatase 1, regulatory inhibitor subunit 14C |
| chr15_-_100976370 | 0.10 |
ENSMUST00000213610.2
|
Fignl2
|
fidgetin-like 2 |
| chr19_+_37538843 | 0.10 |
ENSMUST00000066439.8
ENSMUST00000238817.2 |
Exoc6
|
exocyst complex component 6 |
| chr5_-_137608886 | 0.10 |
ENSMUST00000142675.8
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr19_+_10611574 | 0.10 |
ENSMUST00000055115.9
|
Vwce
|
von Willebrand factor C and EGF domains |
| chr10_-_79938487 | 0.10 |
ENSMUST00000042771.8
|
Sbno2
|
strawberry notch 2 |
| chr2_+_121697398 | 0.10 |
ENSMUST00000110586.10
ENSMUST00000078752.10 |
Golm2
|
golgi membrane protein 2 |
| chr5_+_110324506 | 0.10 |
ENSMUST00000112512.8
|
Golga3
|
golgi autoantigen, golgin subfamily a, 3 |
| chr12_-_100691316 | 0.10 |
ENSMUST00000222731.2
|
Rps6ka5
|
ribosomal protein S6 kinase, polypeptide 5 |
| chr2_-_25517945 | 0.10 |
ENSMUST00000028307.9
|
Fcna
|
ficolin A |
| chr7_-_102368277 | 0.10 |
ENSMUST00000216312.2
|
Olfr33
|
olfactory receptor 33 |
| chr7_-_28465870 | 0.10 |
ENSMUST00000085851.12
ENSMUST00000032815.11 |
Nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, beta |
| chr9_-_65815958 | 0.10 |
ENSMUST00000119245.8
ENSMUST00000134338.8 ENSMUST00000179395.8 |
Trip4
|
thyroid hormone receptor interactor 4 |
| chr2_-_146353819 | 0.10 |
ENSMUST00000131824.8
|
Ralgapa2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr2_+_136555364 | 0.10 |
ENSMUST00000028727.11
ENSMUST00000110098.4 |
Snap25
|
synaptosomal-associated protein 25 |
| chr12_+_87194476 | 0.09 |
ENSMUST00000063117.10
ENSMUST00000220574.2 |
Gstz1
|
glutathione transferase zeta 1 (maleylacetoacetate isomerase) |
| chr11_+_120612369 | 0.09 |
ENSMUST00000142229.2
|
Rac3
|
Rac family small GTPase 3 |
| chr15_-_79287747 | 0.09 |
ENSMUST00000074991.10
|
Tmem184b
|
transmembrane protein 184b |
| chr19_-_47452557 | 0.09 |
ENSMUST00000111800.4
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr3_-_106455042 | 0.09 |
ENSMUST00000039153.14
ENSMUST00000148269.2 ENSMUST00000068301.11 |
Cept1
|
choline/ethanolaminephosphotransferase 1 |
| chr1_-_171188302 | 0.09 |
ENSMUST00000061878.5
|
Klhdc9
|
kelch domain containing 9 |
| chr18_-_35836161 | 0.09 |
ENSMUST00000025208.7
|
Dnajc18
|
DnaJ heat shock protein family (Hsp40) member C18 |
| chr5_+_73071897 | 0.09 |
ENSMUST00000200785.2
|
Slain2
|
SLAIN motif family, member 2 |
| chr16_+_33504829 | 0.09 |
ENSMUST00000152782.8
|
Heg1
|
heart development protein with EGF-like domains 1 |
| chr10_+_79690492 | 0.09 |
ENSMUST00000171599.8
ENSMUST00000095457.11 |
Ptbp1
|
polypyrimidine tract binding protein 1 |
| chr12_+_112688597 | 0.09 |
ENSMUST00000101018.11
ENSMUST00000092279.7 ENSMUST00000179041.8 ENSMUST00000222711.2 |
Cep170b
|
centrosomal protein 170B |
| chr5_-_107437427 | 0.09 |
ENSMUST00000031224.15
|
Tgfbr3
|
transforming growth factor, beta receptor III |
| chr4_-_135601098 | 0.09 |
ENSMUST00000030436.12
|
Pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr9_-_107749886 | 0.08 |
ENSMUST00000195883.6
|
Rbm6
|
RNA binding motif protein 6 |
| chr12_-_111344038 | 0.08 |
ENSMUST00000222196.2
|
Cdc42bpb
|
CDC42 binding protein kinase beta |
| chr7_-_24705320 | 0.08 |
ENSMUST00000102858.10
ENSMUST00000196684.2 ENSMUST00000080882.11 |
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr9_-_21982637 | 0.08 |
ENSMUST00000179605.9
ENSMUST00000043922.7 |
Zfp653
|
zinc finger protein 653 |
| chr7_+_44667377 | 0.08 |
ENSMUST00000044111.10
|
Rras
|
related RAS viral (r-ras) oncogene |
| chr2_-_120680870 | 0.08 |
ENSMUST00000143051.8
ENSMUST00000057135.14 ENSMUST00000085840.11 |
Ttbk2
|
tau tubulin kinase 2 |
| chr4_-_148711211 | 0.08 |
ENSMUST00000186947.7
ENSMUST00000105699.8 |
Tardbp
|
TAR DNA binding protein |
| chr9_+_100956145 | 0.08 |
ENSMUST00000189616.2
|
Msl2
|
MSL complex subunit 2 |
| chr11_+_74540284 | 0.08 |
ENSMUST00000117818.2
ENSMUST00000092915.12 |
Cluh
|
clustered mitochondria (cluA/CLU1) homolog |
| chr15_-_79287447 | 0.08 |
ENSMUST00000228002.2
|
Tmem184b
|
transmembrane protein 184b |
| chr15_-_75886166 | 0.07 |
ENSMUST00000060807.12
|
Fam83h
|
family with sequence similarity 83, member H |
| chr9_-_107749982 | 0.07 |
ENSMUST00000183032.8
ENSMUST00000035201.13 |
Rbm6
|
RNA binding motif protein 6 |
| chr4_+_136038243 | 0.07 |
ENSMUST00000131671.8
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr12_+_71183622 | 0.07 |
ENSMUST00000149564.8
|
2700049A03Rik
|
RIKEN cDNA 2700049A03 gene |
| chr7_-_66339319 | 0.07 |
ENSMUST00000124899.8
|
Asb7
|
ankyrin repeat and SOCS box-containing 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp148
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1900191 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.1 | 0.8 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.1 | 0.3 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.1 | 0.3 | GO:0002353 | plasma kallikrein-kinin cascade(GO:0002353) |
| 0.1 | 0.3 | GO:2000793 | cell proliferation involved in heart valve development(GO:2000793) |
| 0.1 | 0.7 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.4 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.1 | 0.7 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.3 | GO:0003017 | lymph circulation(GO:0003017) |
| 0.1 | 0.2 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.1 | 0.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.3 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.3 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.2 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.2 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.7 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.2 | GO:0040010 | positive regulation of growth rate(GO:0040010) |
| 0.0 | 0.1 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.0 | 0.3 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.0 | 0.4 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0000239 | pachytene(GO:0000239) |
| 0.0 | 0.1 | GO:0060935 | transforming growth factor beta receptor complex assembly(GO:0007181) cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) |
| 0.0 | 0.1 | GO:1990164 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.7 | GO:1905144 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.3 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0061076 | negative regulation of neural retina development(GO:0061076) negative regulation of retina development in camera-type eye(GO:1902867) negative regulation of amacrine cell differentiation(GO:1902870) |
| 0.0 | 0.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.4 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.1 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.4 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.1 | GO:1904414 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.0 | 0.4 | GO:0045060 | negative thymic T cell selection(GO:0045060) |
| 0.0 | 0.1 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.2 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.3 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.0 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 0.0 | 0.1 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.0 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 0.0 | 0.1 | GO:1902474 | regulation of cellular response to heat(GO:1900034) positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 1.3 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.3 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.2 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.2 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.0 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.0 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.2 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.2 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:0042939 | regulation of sodium:proton antiporter activity(GO:0032415) glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.1 | 0.5 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.2 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.2 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.7 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.0 | 0.0 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.2 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.1 | 0.4 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.2 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.1 | 0.2 | GO:0016034 | maleylacetoacetate isomerase activity(GO:0016034) |
| 0.1 | 0.4 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0019002 | GMP binding(GO:0019002) |
| 0.0 | 0.2 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.1 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 1.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.2 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.5 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.0 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.0 | GO:0045159 | myosin II binding(GO:0045159) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.5 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 1.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.3 | PID S1P S1P2 PATHWAY | S1P2 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 1.0 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |