Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Zfp219_Zfp740
Z-value: 0.79
Transcription factors associated with Zfp219_Zfp740
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
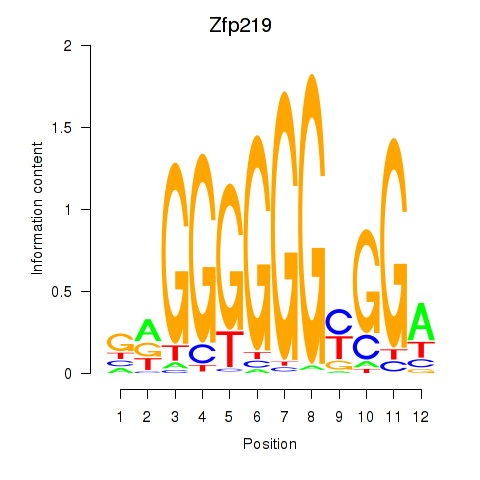
Zfp219
|
ENSMUSG00000049295.18 | zinc finger protein 219 |
|
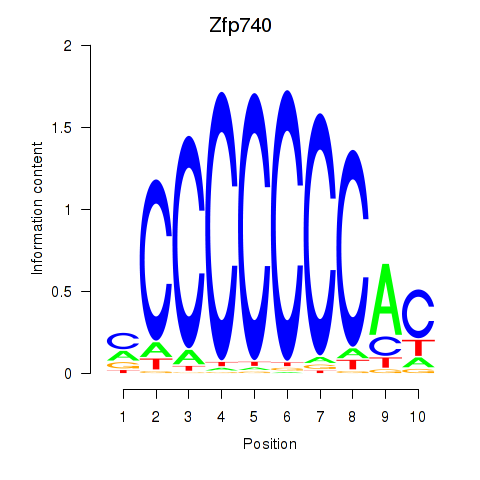
Zfp740
|
ENSMUSG00000046897.18 | zinc finger protein 740 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp219 | mm39_v1_chr14_-_52252318_52252369 | 0.95 | 1.4e-02 | Click! |
| Zfp740 | mm39_v1_chr15_+_102112074_102112144 | 0.90 | 3.8e-02 | Click! |
Activity profile of Zfp219_Zfp740 motif
Sorted Z-values of Zfp219_Zfp740 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_34124078 | 0.64 |
ENSMUST00000172817.2
|
Smim40
|
small integral membrane protein 40 |
| chr3_+_41519289 | 0.37 |
ENSMUST00000168086.7
|
Jade1
|
jade family PHD finger 1 |
| chr8_-_85413707 | 0.35 |
ENSMUST00000238301.2
|
Nacc1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr17_-_28039506 | 0.31 |
ENSMUST00000114859.9
ENSMUST00000233533.2 |
Ilrun
|
inflammation and lipid regulator with UBA-like and NBR1-like domains |
| chr10_-_37014859 | 0.31 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr2_+_116951855 | 0.30 |
ENSMUST00000028829.13
|
Spred1
|
sprouty protein with EVH-1 domain 1, related sequence |
| chr4_-_133746138 | 0.29 |
ENSMUST00000051674.3
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chr11_+_98686321 | 0.29 |
ENSMUST00000037915.9
|
Msl1
|
male specific lethal 1 |
| chr7_+_4693759 | 0.29 |
ENSMUST00000048248.9
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr4_-_87951565 | 0.27 |
ENSMUST00000078090.12
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr7_+_4693603 | 0.27 |
ENSMUST00000120836.8
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr3_+_41519355 | 0.25 |
ENSMUST00000191952.2
|
Jade1
|
jade family PHD finger 1 |
| chr17_-_28039588 | 0.24 |
ENSMUST00000114863.10
ENSMUST00000233131.2 |
Ilrun
|
inflammation and lipid regulator with UBA-like and NBR1-like domains |
| chr1_-_72914036 | 0.23 |
ENSMUST00000027377.9
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr7_-_68398989 | 0.22 |
ENSMUST00000048068.15
|
Arrdc4
|
arrestin domain containing 4 |
| chr11_-_102187445 | 0.22 |
ENSMUST00000107132.3
ENSMUST00000073234.9 |
Atxn7l3
|
ataxin 7-like 3 |
| chr7_+_127345909 | 0.21 |
ENSMUST00000033081.14
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr6_+_51447613 | 0.20 |
ENSMUST00000114445.8
ENSMUST00000114446.8 ENSMUST00000141711.3 |
Cbx3
|
chromobox 3 |
| chr10_+_43455157 | 0.20 |
ENSMUST00000058714.10
|
Cd24a
|
CD24a antigen |
| chr4_-_3938352 | 0.20 |
ENSMUST00000003369.10
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chr7_+_90091937 | 0.19 |
ENSMUST00000061767.5
|
Crebzf
|
CREB/ATF bZIP transcription factor |
| chr9_+_72439891 | 0.19 |
ENSMUST00000183372.8
ENSMUST00000184015.2 |
Rfx7
|
regulatory factor X, 7 |
| chr2_+_156681991 | 0.18 |
ENSMUST00000073352.10
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chrX_+_100298134 | 0.18 |
ENSMUST00000062000.6
|
Foxo4
|
forkhead box O4 |
| chr2_-_48839218 | 0.17 |
ENSMUST00000090976.10
ENSMUST00000149679.8 |
Orc4
|
origin recognition complex, subunit 4 |
| chr9_+_14187597 | 0.17 |
ENSMUST00000208222.2
|
Sesn3
|
sestrin 3 |
| chr17_+_35191661 | 0.17 |
ENSMUST00000007248.5
|
Hspa1l
|
heat shock protein 1-like |
| chr3_-_89152320 | 0.17 |
ENSMUST00000107464.8
ENSMUST00000090924.13 |
Trim46
|
tripartite motif-containing 46 |
| chr13_-_30168374 | 0.17 |
ENSMUST00000221536.2
ENSMUST00000222730.2 |
E2f3
|
E2F transcription factor 3 |
| chr18_+_34910064 | 0.16 |
ENSMUST00000043775.9
ENSMUST00000224715.2 |
Kdm3b
|
KDM3B lysine (K)-specific demethylase 3B |
| chr10_+_126914755 | 0.16 |
ENSMUST00000039259.7
ENSMUST00000217941.2 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr12_-_80307110 | 0.16 |
ENSMUST00000021554.16
|
Actn1
|
actinin, alpha 1 |
| chr11_+_98686617 | 0.15 |
ENSMUST00000107485.8
ENSMUST00000107487.10 |
Msl1
|
male specific lethal 1 |
| chr11_+_29642937 | 0.15 |
ENSMUST00000102843.10
ENSMUST00000102842.10 ENSMUST00000078830.11 ENSMUST00000170731.8 |
Rtn4
|
reticulon 4 |
| chr11_-_96714813 | 0.15 |
ENSMUST00000142065.2
ENSMUST00000167110.8 |
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr7_-_81584081 | 0.15 |
ENSMUST00000026094.6
ENSMUST00000107305.8 |
Hdgfl3
|
HDGF like 3 |
| chr7_-_126625739 | 0.15 |
ENSMUST00000205461.2
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr2_-_162502994 | 0.15 |
ENSMUST00000109442.8
ENSMUST00000109445.9 ENSMUST00000109443.8 ENSMUST00000109441.2 |
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chr3_+_129326004 | 0.15 |
ENSMUST00000199910.5
ENSMUST00000197070.5 ENSMUST00000071402.7 |
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr17_+_69690314 | 0.15 |
ENSMUST00000062369.14
|
Zbtb14
|
zinc finger and BTB domain containing 14 |
| chr11_+_69909659 | 0.14 |
ENSMUST00000232002.2
ENSMUST00000134376.10 ENSMUST00000231221.2 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr4_+_32615428 | 0.14 |
ENSMUST00000178925.8
ENSMUST00000029950.10 |
Casp8ap2
|
caspase 8 associated protein 2 |
| chr17_+_35113490 | 0.14 |
ENSMUST00000052778.10
|
Zbtb12
|
zinc finger and BTB domain containing 12 |
| chr18_+_82929451 | 0.14 |
ENSMUST00000235902.2
|
Zfp516
|
zinc finger protein 516 |
| chr14_-_55828511 | 0.14 |
ENSMUST00000161807.8
ENSMUST00000111378.10 ENSMUST00000159687.2 |
Psme2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr17_+_34149820 | 0.14 |
ENSMUST00000234226.2
|
Rgl2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr8_-_85414220 | 0.14 |
ENSMUST00000238449.2
ENSMUST00000238687.2 |
Nacc1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr17_-_28039419 | 0.14 |
ENSMUST00000233508.2
ENSMUST00000075076.6 |
Ilrun
|
inflammation and lipid regulator with UBA-like and NBR1-like domains |
| chr7_-_89629809 | 0.14 |
ENSMUST00000238792.2
|
Eed
|
embryonic ectoderm development |
| chr11_-_102208615 | 0.14 |
ENSMUST00000107117.9
|
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr11_+_94881861 | 0.14 |
ENSMUST00000038696.12
|
Ppp1r9b
|
protein phosphatase 1, regulatory subunit 9B |
| chr7_-_24771717 | 0.14 |
ENSMUST00000003468.10
|
Grik5
|
glutamate receptor, ionotropic, kainate 5 (gamma 2) |
| chr3_-_95125190 | 0.14 |
ENSMUST00000136139.8
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr12_-_80306865 | 0.13 |
ENSMUST00000167327.2
|
Actn1
|
actinin, alpha 1 |
| chr11_-_102208449 | 0.13 |
ENSMUST00000107123.10
|
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr7_-_126303689 | 0.13 |
ENSMUST00000135087.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr1_-_84817000 | 0.13 |
ENSMUST00000186648.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr2_-_153286361 | 0.13 |
ENSMUST00000109784.2
|
Nol4l
|
nucleolar protein 4-like |
| chr4_-_118037143 | 0.13 |
ENSMUST00000050288.9
ENSMUST00000106403.8 |
Kdm4a
|
lysine (K)-specific demethylase 4A |
| chr11_+_23206001 | 0.12 |
ENSMUST00000020538.13
ENSMUST00000109551.8 ENSMUST00000102870.8 ENSMUST00000102869.8 |
Xpo1
|
exportin 1 |
| chr5_+_32616566 | 0.12 |
ENSMUST00000202078.2
|
Ppp1cb
|
protein phosphatase 1 catalytic subunit beta |
| chr11_+_3239868 | 0.12 |
ENSMUST00000094471.10
ENSMUST00000110043.8 |
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr7_-_25488060 | 0.12 |
ENSMUST00000002677.11
ENSMUST00000085948.11 |
Axl
|
AXL receptor tyrosine kinase |
| chr4_-_118036827 | 0.11 |
ENSMUST00000097911.9
|
Kdm4a
|
lysine (K)-specific demethylase 4A |
| chr2_-_48839276 | 0.11 |
ENSMUST00000028098.11
|
Orc4
|
origin recognition complex, subunit 4 |
| chr18_-_56695333 | 0.11 |
ENSMUST00000066208.13
ENSMUST00000172734.8 |
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr4_-_136563154 | 0.11 |
ENSMUST00000105846.9
ENSMUST00000059287.14 ENSMUST00000105845.9 |
Ephb2
|
Eph receptor B2 |
| chr12_-_118265103 | 0.11 |
ENSMUST00000222314.2
ENSMUST00000026367.11 |
Sp4
|
trans-acting transcription factor 4 |
| chr9_+_35179153 | 0.11 |
ENSMUST00000034543.5
|
Rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr4_+_129083553 | 0.11 |
ENSMUST00000106054.4
|
Yars
|
tyrosyl-tRNA synthetase |
| chr17_-_47169380 | 0.11 |
ENSMUST00000233455.2
|
Bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr18_+_35962832 | 0.11 |
ENSMUST00000060722.8
|
Cxxc5
|
CXXC finger 5 |
| chr11_-_69872050 | 0.11 |
ENSMUST00000108594.8
|
Elp5
|
elongator acetyltransferase complex subunit 5 |
| chr3_-_95125002 | 0.11 |
ENSMUST00000107209.8
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr12_-_73160181 | 0.11 |
ENSMUST00000043208.8
ENSMUST00000175693.3 |
Six4
|
sine oculis-related homeobox 4 |
| chr16_+_10884156 | 0.11 |
ENSMUST00000089011.6
|
Snn
|
stannin |
| chr10_+_11157047 | 0.11 |
ENSMUST00000129456.8
|
Fbxo30
|
F-box protein 30 |
| chr11_-_87878301 | 0.11 |
ENSMUST00000020775.9
|
Dynll2
|
dynein light chain LC8-type 2 |
| chr8_+_112263632 | 0.10 |
ENSMUST00000173506.8
|
Znrf1
|
zinc and ring finger 1 |
| chr7_+_127376267 | 0.10 |
ENSMUST00000144406.8
|
Setd1a
|
SET domain containing 1A |
| chr11_+_69792642 | 0.10 |
ENSMUST00000177138.8
ENSMUST00000108617.10 ENSMUST00000177476.8 ENSMUST00000061837.11 |
Neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr4_-_148711453 | 0.10 |
ENSMUST00000165113.8
ENSMUST00000172073.8 ENSMUST00000105702.9 ENSMUST00000084125.10 |
Tardbp
|
TAR DNA binding protein |
| chr16_+_20492267 | 0.10 |
ENSMUST00000115460.8
|
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr7_-_126625657 | 0.10 |
ENSMUST00000205568.2
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr16_+_20536856 | 0.10 |
ENSMUST00000231392.2
ENSMUST00000161038.2 |
Polr2h
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr4_+_97665843 | 0.10 |
ENSMUST00000075448.13
ENSMUST00000092532.13 |
Nfia
|
nuclear factor I/A |
| chr18_+_69478790 | 0.10 |
ENSMUST00000202116.4
ENSMUST00000114982.8 ENSMUST00000078486.13 ENSMUST00000202772.4 ENSMUST00000201288.4 |
Tcf4
|
transcription factor 4 |
| chr2_-_84605732 | 0.10 |
ENSMUST00000023994.10
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr15_+_102379503 | 0.09 |
ENSMUST00000229222.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr7_-_89630141 | 0.09 |
ENSMUST00000238981.2
ENSMUST00000208977.2 ENSMUST00000107234.3 |
Eed
|
embryonic ectoderm development |
| chr10_-_93146825 | 0.09 |
ENSMUST00000151153.2
|
Elk3
|
ELK3, member of ETS oncogene family |
| chr9_+_21108001 | 0.09 |
ENSMUST00000003395.10
|
Pde4a
|
phosphodiesterase 4A, cAMP specific |
| chr17_-_46558894 | 0.09 |
ENSMUST00000142706.9
ENSMUST00000173349.8 ENSMUST00000087026.13 |
Polr1c
|
polymerase (RNA) I polypeptide C |
| chr17_+_34148868 | 0.09 |
ENSMUST00000173266.8
|
Rgl2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr4_-_118037240 | 0.09 |
ENSMUST00000106406.9
|
Kdm4a
|
lysine (K)-specific demethylase 4A |
| chr13_+_55359598 | 0.09 |
ENSMUST00000224918.2
|
Nsd1
|
nuclear receptor-binding SET-domain protein 1 |
| chr12_-_112893382 | 0.09 |
ENSMUST00000075827.5
|
Jag2
|
jagged 2 |
| chr9_-_20888054 | 0.09 |
ENSMUST00000054197.7
|
S1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr11_+_23256883 | 0.09 |
ENSMUST00000180046.8
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr11_-_97635484 | 0.09 |
ENSMUST00000018691.9
|
Pip4k2b
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta |
| chr11_-_96714950 | 0.09 |
ENSMUST00000169828.8
ENSMUST00000126949.8 |
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr3_+_104545974 | 0.09 |
ENSMUST00000046212.2
|
Slc16a1
|
solute carrier family 16 (monocarboxylic acid transporters), member 1 |
| chr19_+_46611826 | 0.09 |
ENSMUST00000111855.5
|
Wbp1l
|
WW domain binding protein 1 like |
| chr2_+_156681927 | 0.09 |
ENSMUST00000081335.13
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr8_+_40876827 | 0.09 |
ENSMUST00000049389.11
ENSMUST00000128166.8 ENSMUST00000167766.2 |
Zdhhc2
|
zinc finger, DHHC domain containing 2 |
| chr7_+_89779493 | 0.09 |
ENSMUST00000208730.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr11_+_3845221 | 0.09 |
ENSMUST00000109996.8
ENSMUST00000055931.5 |
Dusp18
|
dual specificity phosphatase 18 |
| chr2_+_4887015 | 0.09 |
ENSMUST00000115019.2
|
Sephs1
|
selenophosphate synthetase 1 |
| chrX_+_150127171 | 0.09 |
ENSMUST00000073364.6
|
Fam120c
|
family with sequence similarity 120, member C |
| chr3_+_95836558 | 0.09 |
ENSMUST00000165307.8
ENSMUST00000015893.13 ENSMUST00000169426.8 |
Anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chrX_-_98514278 | 0.08 |
ENSMUST00000113797.4
ENSMUST00000113790.8 ENSMUST00000036354.7 ENSMUST00000167246.2 |
Pja1
|
praja ring finger ubiquitin ligase 1 |
| chr10_+_11157326 | 0.08 |
ENSMUST00000070300.5
|
Fbxo30
|
F-box protein 30 |
| chr11_-_98040377 | 0.08 |
ENSMUST00000103143.10
|
Fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr5_+_118698689 | 0.08 |
ENSMUST00000100816.8
ENSMUST00000201010.2 |
Med13l
|
mediator complex subunit 13-like |
| chrX_+_37689503 | 0.08 |
ENSMUST00000000365.3
|
Mcts1
|
malignant T cell amplified sequence 1 |
| chr3_-_104725581 | 0.08 |
ENSMUST00000168015.8
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr8_-_105169621 | 0.08 |
ENSMUST00000041769.8
|
Dync1li2
|
dynein, cytoplasmic 1 light intermediate chain 2 |
| chrX_+_7688528 | 0.08 |
ENSMUST00000009875.5
|
Kcnd1
|
potassium voltage-gated channel, Shal-related family, member 1 |
| chr11_+_3599183 | 0.08 |
ENSMUST00000096441.5
|
Morc2a
|
microrchidia 2A |
| chr11_+_115310963 | 0.08 |
ENSMUST00000106533.8
ENSMUST00000123345.2 |
Kctd2
|
potassium channel tetramerisation domain containing 2 |
| chr11_-_69812016 | 0.08 |
ENSMUST00000108607.8
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr1_+_93731662 | 0.08 |
ENSMUST00000027505.13
|
Ing5
|
inhibitor of growth family, member 5 |
| chr17_-_56447332 | 0.08 |
ENSMUST00000001256.11
|
Sema6b
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr12_+_87194476 | 0.08 |
ENSMUST00000063117.10
ENSMUST00000220574.2 |
Gstz1
|
glutathione transferase zeta 1 (maleylacetoacetate isomerase) |
| chrX_-_93166992 | 0.08 |
ENSMUST00000088102.12
ENSMUST00000113927.8 |
Zfx
|
zinc finger protein X-linked |
| chr11_+_115310885 | 0.08 |
ENSMUST00000103035.10
|
Kctd2
|
potassium channel tetramerisation domain containing 2 |
| chr14_-_71003973 | 0.08 |
ENSMUST00000226448.2
ENSMUST00000022696.8 |
Xpo7
|
exportin 7 |
| chr3_+_129326285 | 0.08 |
ENSMUST00000197235.5
|
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr9_+_31191820 | 0.08 |
ENSMUST00000117389.8
ENSMUST00000215499.2 |
Prdm10
|
PR domain containing 10 |
| chrX_-_35909011 | 0.08 |
ENSMUST00000051906.13
|
Akap17b
|
A kinase (PRKA) anchor protein 17B |
| chr7_+_89779421 | 0.08 |
ENSMUST00000207225.2
ENSMUST00000207484.2 ENSMUST00000209068.2 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr18_-_10030017 | 0.08 |
ENSMUST00000116669.2
ENSMUST00000092096.14 |
Usp14
|
ubiquitin specific peptidase 14 |
| chr1_+_156386414 | 0.08 |
ENSMUST00000166172.9
ENSMUST00000027888.13 |
Abl2
|
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
| chr15_+_102427149 | 0.08 |
ENSMUST00000146756.8
ENSMUST00000142194.3 |
Tarbp2
|
TARBP2, RISC loading complex RNA binding subunit |
| chr4_-_133399909 | 0.08 |
ENSMUST00000062118.11
ENSMUST00000067902.13 |
Pigv
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr12_+_87193922 | 0.08 |
ENSMUST00000222885.2
|
Gstz1
|
glutathione transferase zeta 1 (maleylacetoacetate isomerase) |
| chr12_+_85646162 | 0.08 |
ENSMUST00000050687.14
|
Jdp2
|
Jun dimerization protein 2 |
| chr3_-_94566107 | 0.07 |
ENSMUST00000196655.5
ENSMUST00000200407.2 ENSMUST00000006123.11 ENSMUST00000196733.5 |
Tuft1
|
tuftelin 1 |
| chr4_+_98284128 | 0.07 |
ENSMUST00000107030.9
|
Patj
|
PATJ, crumbs cell polarity complex component |
| chr15_+_82031382 | 0.07 |
ENSMUST00000023100.8
ENSMUST00000229336.2 |
Srebf2
|
sterol regulatory element binding factor 2 |
| chr16_-_38533597 | 0.07 |
ENSMUST00000023487.5
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr12_+_4183394 | 0.07 |
ENSMUST00000152065.8
ENSMUST00000127756.8 |
Adcy3
|
adenylate cyclase 3 |
| chr13_+_24986003 | 0.07 |
ENSMUST00000155575.2
|
BC005537
|
cDNA sequence BC005537 |
| chr15_-_89310060 | 0.07 |
ENSMUST00000109313.9
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr17_+_35117905 | 0.07 |
ENSMUST00000097342.10
ENSMUST00000013931.12 |
Ehmt2
|
euchromatic histone lysine N-methyltransferase 2 |
| chr8_+_85696396 | 0.07 |
ENSMUST00000109733.8
|
Prdx2
|
peroxiredoxin 2 |
| chr5_-_24597009 | 0.07 |
ENSMUST00000059401.7
|
Atg9b
|
autophagy related 9B |
| chr9_+_65253374 | 0.07 |
ENSMUST00000048184.4
ENSMUST00000214433.2 |
Pdcd7
|
programmed cell death 7 |
| chr11_-_6015538 | 0.07 |
ENSMUST00000101585.10
ENSMUST00000066431.14 ENSMUST00000109815.9 ENSMUST00000109812.9 ENSMUST00000101586.3 ENSMUST00000093355.12 ENSMUST00000019133.11 |
Camk2b
|
calcium/calmodulin-dependent protein kinase II, beta |
| chr1_-_168259465 | 0.07 |
ENSMUST00000176540.8
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr17_+_34135145 | 0.07 |
ENSMUST00000053429.11
ENSMUST00000234582.2 |
Zbtb22
Tapbp
|
zinc finger and BTB domain containing 22 TAP binding protein |
| chr7_-_126625617 | 0.07 |
ENSMUST00000032916.6
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr8_+_112262729 | 0.07 |
ENSMUST00000172856.8
|
Znrf1
|
zinc and ring finger 1 |
| chr8_+_112263465 | 0.07 |
ENSMUST00000095176.12
|
Znrf1
|
zinc and ring finger 1 |
| chr9_-_45895635 | 0.07 |
ENSMUST00000215427.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr3_+_20111958 | 0.07 |
ENSMUST00000002502.12
|
Hltf
|
helicase-like transcription factor |
| chr5_-_137529465 | 0.07 |
ENSMUST00000150063.9
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr16_-_13977084 | 0.07 |
ENSMUST00000090300.6
|
Marf1
|
meiosis regulator and mRNA stability 1 |
| chr11_+_69909245 | 0.07 |
ENSMUST00000231415.2
ENSMUST00000108588.9 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr16_+_20536545 | 0.07 |
ENSMUST00000231656.2
|
Polr2h
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr8_+_85696695 | 0.07 |
ENSMUST00000164807.2
|
Prdx2
|
peroxiredoxin 2 |
| chr5_+_3853160 | 0.07 |
ENSMUST00000080085.9
ENSMUST00000200386.5 |
Krit1
|
KRIT1, ankyrin repeat containing |
| chr17_+_27160356 | 0.07 |
ENSMUST00000229490.2
ENSMUST00000201702.5 ENSMUST00000177932.7 ENSMUST00000201349.6 |
Syngap1
|
synaptic Ras GTPase activating protein 1 homolog (rat) |
| chr3_-_104725535 | 0.07 |
ENSMUST00000002297.12
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr11_-_120881904 | 0.07 |
ENSMUST00000070575.14
|
Csnk1d
|
casein kinase 1, delta |
| chr14_+_55120777 | 0.07 |
ENSMUST00000022806.10
|
Bcl2l2
|
BCL2-like 2 |
| chr11_-_88608920 | 0.07 |
ENSMUST00000092794.12
|
Msi2
|
musashi RNA-binding protein 2 |
| chr2_-_34803988 | 0.06 |
ENSMUST00000028232.7
ENSMUST00000202907.2 |
Phf19
|
PHD finger protein 19 |
| chr1_-_164285914 | 0.06 |
ENSMUST00000027863.13
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr18_+_73706115 | 0.06 |
ENSMUST00000091852.5
|
Mex3c
|
mex3 RNA binding family member C |
| chr17_+_69690018 | 0.06 |
ENSMUST00000112674.8
|
Zbtb14
|
zinc finger and BTB domain containing 14 |
| chr2_-_121101822 | 0.06 |
ENSMUST00000110647.8
ENSMUST00000110648.8 |
Trp53bp1
|
transformation related protein 53 binding protein 1 |
| chr3_-_95125051 | 0.06 |
ENSMUST00000107204.8
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr9_-_35179042 | 0.06 |
ENSMUST00000217306.2
ENSMUST00000125087.2 ENSMUST00000121564.8 ENSMUST00000063782.12 ENSMUST00000059057.14 |
Fam118b
|
family with sequence similarity 118, member B |
| chr19_+_6952580 | 0.06 |
ENSMUST00000237084.2
ENSMUST00000236218.2 ENSMUST00000237235.2 |
Ppp1r14b
|
protein phosphatase 1, regulatory inhibitor subunit 14B |
| chr12_+_3941728 | 0.06 |
ENSMUST00000172689.8
ENSMUST00000111186.8 |
Dnmt3a
|
DNA methyltransferase 3A |
| chr8_+_110432132 | 0.06 |
ENSMUST00000212964.2
ENSMUST00000034163.9 |
Zfp821
|
zinc finger protein 821 |
| chr10_+_79986988 | 0.06 |
ENSMUST00000146516.8
ENSMUST00000144526.2 |
Midn
|
midnolin |
| chr16_-_10603389 | 0.06 |
ENSMUST00000229866.2
ENSMUST00000038099.6 |
Socs1
|
suppressor of cytokine signaling 1 |
| chr17_+_85264134 | 0.06 |
ENSMUST00000112305.10
|
Ppm1b
|
protein phosphatase 1B, magnesium dependent, beta isoform |
| chr6_+_83162928 | 0.06 |
ENSMUST00000113907.2
|
Dctn1
|
dynactin 1 |
| chr11_-_95956176 | 0.06 |
ENSMUST00000100528.5
|
Ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr8_+_85696453 | 0.06 |
ENSMUST00000125893.8
|
Prdx2
|
peroxiredoxin 2 |
| chr11_+_23206565 | 0.06 |
ENSMUST00000136235.2
|
Xpo1
|
exportin 1 |
| chr17_+_69690764 | 0.06 |
ENSMUST00000112676.4
|
Zbtb14
|
zinc finger and BTB domain containing 14 |
| chr8_-_106015682 | 0.06 |
ENSMUST00000212922.2
ENSMUST00000212219.2 |
4931428F04Rik
|
RIKEN cDNA 4931428F04 gene |
| chr2_+_84564394 | 0.06 |
ENSMUST00000238573.2
ENSMUST00000090729.9 |
Ypel4
|
yippee like 4 |
| chr10_+_80765900 | 0.06 |
ENSMUST00000015456.10
ENSMUST00000220246.2 |
Gadd45b
|
growth arrest and DNA-damage-inducible 45 beta |
| chr11_+_106107752 | 0.06 |
ENSMUST00000021046.6
|
Ddx42
|
DEAD box helicase 42 |
| chr3_+_86131970 | 0.06 |
ENSMUST00000192145.6
ENSMUST00000194759.6 ENSMUST00000107635.7 |
Lrba
|
LPS-responsive beige-like anchor |
| chr8_-_58106057 | 0.06 |
ENSMUST00000034021.12
|
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr10_-_127124718 | 0.06 |
ENSMUST00000156208.2
ENSMUST00000026476.13 |
Mbd6
|
methyl-CpG binding domain protein 6 |
| chr8_+_85696216 | 0.06 |
ENSMUST00000109734.8
ENSMUST00000005292.15 |
Prdx2
|
peroxiredoxin 2 |
| chrX_+_108138965 | 0.06 |
ENSMUST00000033598.9
|
Sh3bgrl
|
SH3-binding domain glutamic acid-rich protein like |
| chr11_-_74787877 | 0.06 |
ENSMUST00000057631.12
ENSMUST00000081799.6 |
Sgsm2
|
small G protein signaling modulator 2 |
| chr11_+_20493306 | 0.05 |
ENSMUST00000093292.11
|
Sertad2
|
SERTA domain containing 2 |
| chr4_-_148711211 | 0.05 |
ENSMUST00000186947.7
ENSMUST00000105699.8 |
Tardbp
|
TAR DNA binding protein |
| chr11_+_97253221 | 0.05 |
ENSMUST00000238729.2
ENSMUST00000045540.4 |
Socs7
|
suppressor of cytokine signaling 7 |
| chr2_-_160950936 | 0.05 |
ENSMUST00000039782.14
ENSMUST00000134178.8 |
Chd6
|
chromodomain helicase DNA binding protein 6 |
| chr17_+_35220252 | 0.05 |
ENSMUST00000174260.8
|
Vars
|
valyl-tRNA synthetase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp219_Zfp740
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:2000011 | adaxial/abaxial pattern specification(GO:0009955) regulation of adaxial/abaxial pattern formation(GO:2000011) |
| 0.1 | 0.2 | GO:0034117 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 0.1 | 0.2 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0071317 | cellular response to isoquinoline alkaloid(GO:0071317) regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 | 0.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.0 | 0.2 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.1 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.0 | 0.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.3 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.4 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.3 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.1 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0060466 | activation of meiosis involved in egg activation(GO:0060466) |
| 0.0 | 0.1 | GO:0097048 | dendritic cell apoptotic process(GO:0097048) neutrophil clearance(GO:0097350) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.0 | 0.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.0 | 0.3 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.0 | 0.2 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.0 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.3 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 | 0.0 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.2 | GO:0098970 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 | 0.1 | GO:0072368 | regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.1 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:0050689 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) siRNA loading onto RISC involved in RNA interference(GO:0035087) negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.1 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.1 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.1 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.6 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.1 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.1 | 0.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 0.3 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.2 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.0 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.0 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0016034 | maleylacetoacetate isomerase activity(GO:0016034) |
| 0.0 | 0.3 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.1 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.4 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.0 | 0.2 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.0 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |