Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
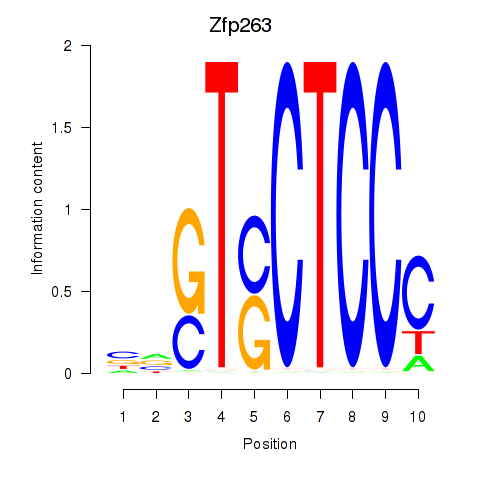
Results for Zfp263
Z-value: 0.89
Transcription factors associated with Zfp263
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp263
|
ENSMUSG00000022529.12 | zinc finger protein 263 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp263 | mm39_v1_chr16_+_3561952_3562023 | 0.20 | 7.5e-01 | Click! |
Activity profile of Zfp263 motif
Sorted Z-values of Zfp263 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_71318353 | 0.71 |
ENSMUST00000064780.4
|
Gabre
|
gamma-aminobutyric acid (GABA) A receptor, subunit epsilon |
| chr11_+_98851238 | 0.69 |
ENSMUST00000107473.3
|
Rara
|
retinoic acid receptor, alpha |
| chr7_-_127423641 | 0.59 |
ENSMUST00000106267.5
|
Stx1b
|
syntaxin 1B |
| chr5_+_90907207 | 0.57 |
ENSMUST00000031318.6
|
Cxcl5
|
chemokine (C-X-C motif) ligand 5 |
| chr11_+_16702203 | 0.48 |
ENSMUST00000102884.10
ENSMUST00000020329.13 |
Egfr
|
epidermal growth factor receptor |
| chr5_-_25703700 | 0.46 |
ENSMUST00000173073.8
ENSMUST00000045291.14 ENSMUST00000173174.2 |
Kmt2c
|
lysine (K)-specific methyltransferase 2C |
| chr12_-_99359265 | 0.41 |
ENSMUST00000177451.8
|
Foxn3
|
forkhead box N3 |
| chr1_-_170417354 | 0.40 |
ENSMUST00000160456.8
|
Nos1ap
|
nitric oxide synthase 1 (neuronal) adaptor protein |
| chr8_-_107064615 | 0.39 |
ENSMUST00000067512.8
|
Smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr7_+_100355910 | 0.39 |
ENSMUST00000207875.2
ENSMUST00000208013.2 |
Fam168a
|
family with sequence similarity 168, member A |
| chr15_+_101310283 | 0.37 |
ENSMUST00000068904.9
|
Krt7
|
keratin 7 |
| chrX_-_20157966 | 0.35 |
ENSMUST00000115393.3
ENSMUST00000072451.11 |
Slc9a7
|
solute carrier family 9 (sodium/hydrogen exchanger), member 7 |
| chr1_-_173569301 | 0.35 |
ENSMUST00000042610.15
ENSMUST00000127730.2 |
Ifi207
|
interferon activated gene 207 |
| chr6_+_6863269 | 0.35 |
ENSMUST00000171311.8
ENSMUST00000160937.9 |
Dlx6
|
distal-less homeobox 6 |
| chrX_-_7242065 | 0.35 |
ENSMUST00000191497.2
ENSMUST00000115744.2 |
Usp27x
|
ubiquitin specific peptidase 27, X chromosome |
| chr12_+_105302853 | 0.33 |
ENSMUST00000180458.9
|
Tunar
|
Tcl1 upstream neural differentiation associated RNA |
| chr6_-_116693849 | 0.32 |
ENSMUST00000056623.13
|
Tmem72
|
transmembrane protein 72 |
| chr7_+_5023552 | 0.31 |
ENSMUST00000208728.2
ENSMUST00000085427.6 |
Ccdc106
Zfp865
|
coiled-coil domain containing 106 zinc finger protein 865 |
| chr7_+_5023375 | 0.31 |
ENSMUST00000076251.7
|
Zfp865
|
zinc finger protein 865 |
| chr2_-_172782089 | 0.30 |
ENSMUST00000009143.8
|
Bmp7
|
bone morphogenetic protein 7 |
| chr11_+_45946800 | 0.30 |
ENSMUST00000011400.8
|
Adam19
|
a disintegrin and metallopeptidase domain 19 (meltrin beta) |
| chr7_-_15781838 | 0.30 |
ENSMUST00000210781.2
|
Bicra
|
BRD4 interacting chromatin remodeling complex associated protein |
| chr4_+_136013372 | 0.30 |
ENSMUST00000069195.5
ENSMUST00000130658.2 |
Zfp46
|
zinc finger protein 46 |
| chr14_-_45456821 | 0.29 |
ENSMUST00000128484.8
ENSMUST00000147853.8 ENSMUST00000022377.11 ENSMUST00000143609.2 ENSMUST00000153383.8 ENSMUST00000139526.9 |
Txndc16
|
thioredoxin domain containing 16 |
| chr13_-_47196633 | 0.28 |
ENSMUST00000021806.11
ENSMUST00000136864.8 |
Tpmt
|
thiopurine methyltransferase |
| chr2_+_54326329 | 0.28 |
ENSMUST00000112636.8
ENSMUST00000112635.8 ENSMUST00000112634.8 |
Galnt13
|
polypeptide N-acetylgalactosaminyltransferase 13 |
| chr2_+_48839505 | 0.28 |
ENSMUST00000112745.8
ENSMUST00000112754.8 |
Mbd5
|
methyl-CpG binding domain protein 5 |
| chr11_-_107685383 | 0.27 |
ENSMUST00000021066.4
|
Cacng4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr7_+_127078371 | 0.26 |
ENSMUST00000205432.3
|
Fbrs
|
fibrosin |
| chrX_+_162691978 | 0.26 |
ENSMUST00000069041.15
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr17_-_47074617 | 0.26 |
ENSMUST00000041012.9
|
Ptcra
|
pre T cell antigen receptor alpha |
| chr16_+_14523696 | 0.25 |
ENSMUST00000023356.8
|
Snai2
|
snail family zinc finger 2 |
| chr5_+_67765216 | 0.25 |
ENSMUST00000087241.7
|
Shisa3
|
shisa family member 3 |
| chr15_-_98769056 | 0.25 |
ENSMUST00000178486.9
ENSMUST00000023741.16 |
Kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr6_-_149003171 | 0.25 |
ENSMUST00000111557.8
|
Dennd5b
|
DENN/MADD domain containing 5B |
| chr7_+_3648264 | 0.24 |
ENSMUST00000206287.2
ENSMUST00000038913.16 |
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr5_-_91550853 | 0.24 |
ENSMUST00000121044.6
|
Btc
|
betacellulin, epidermal growth factor family member |
| chr5_+_135197228 | 0.24 |
ENSMUST00000111187.10
ENSMUST00000111188.5 ENSMUST00000202606.3 |
Bcl7b
|
B cell CLL/lymphoma 7B |
| chr1_-_84262274 | 0.24 |
ENSMUST00000177458.2
ENSMUST00000168574.9 |
Pid1
|
phosphotyrosine interaction domain containing 1 |
| chr5_-_132572181 | 0.24 |
ENSMUST00000161226.11
|
Auts2
|
autism susceptibility candidate 2 |
| chr12_-_84240781 | 0.24 |
ENSMUST00000110294.2
|
Mideas
|
mitotic deacetylase associated SANT domain protein |
| chr10_-_84276454 | 0.23 |
ENSMUST00000020220.15
|
Nuak1
|
NUAK family, SNF1-like kinase, 1 |
| chr16_-_23807602 | 0.23 |
ENSMUST00000023151.6
|
Bcl6
|
B cell leukemia/lymphoma 6 |
| chr1_-_86286690 | 0.23 |
ENSMUST00000185785.2
|
Ncl
|
nucleolin |
| chr7_+_28869770 | 0.22 |
ENSMUST00000033886.8
ENSMUST00000209019.2 ENSMUST00000208330.2 |
Ggn
|
gametogenetin |
| chr9_+_30941924 | 0.22 |
ENSMUST00000216649.2
ENSMUST00000115222.10 |
Zbtb44
|
zinc finger and BTB domain containing 44 |
| chr9_+_109760856 | 0.22 |
ENSMUST00000169851.8
|
Map4
|
microtubule-associated protein 4 |
| chr9_+_102885156 | 0.22 |
ENSMUST00000035148.13
|
Slco2a1
|
solute carrier organic anion transporter family, member 2a1 |
| chr2_-_130748259 | 0.22 |
ENSMUST00000128176.2
ENSMUST00000133766.2 ENSMUST00000135110.2 ENSMUST00000146975.2 |
A730017L22Rik
4930402H24Rik
|
RIKEN cDNA A730017L22 gene RIKEN cDNA 4930402H24 gene |
| chr5_+_150042092 | 0.21 |
ENSMUST00000200960.4
ENSMUST00000202530.4 |
Fry
|
FRY microtubule binding protein |
| chr3_+_86131970 | 0.21 |
ENSMUST00000192145.6
ENSMUST00000194759.6 ENSMUST00000107635.7 |
Lrba
|
LPS-responsive beige-like anchor |
| chr16_-_4376471 | 0.21 |
ENSMUST00000230875.2
|
Tfap4
|
transcription factor AP4 |
| chr6_-_149003003 | 0.21 |
ENSMUST00000127727.2
|
Dennd5b
|
DENN/MADD domain containing 5B |
| chr18_-_15196612 | 0.21 |
ENSMUST00000168989.9
|
Kctd1
|
potassium channel tetramerisation domain containing 1 |
| chr4_+_107825529 | 0.20 |
ENSMUST00000106713.5
ENSMUST00000238795.2 |
Slc1a7
|
solute carrier family 1 (glutamate transporter), member 7 |
| chrX_+_135723420 | 0.20 |
ENSMUST00000033800.13
|
Plp1
|
proteolipid protein (myelin) 1 |
| chrX_-_20787150 | 0.20 |
ENSMUST00000081893.7
ENSMUST00000115345.8 |
Syn1
|
synapsin I |
| chr17_-_74017410 | 0.20 |
ENSMUST00000112591.3
ENSMUST00000024858.12 |
Galnt14
|
polypeptide N-acetylgalactosaminyltransferase 14 |
| chr16_+_35361635 | 0.20 |
ENSMUST00000120756.8
|
Sema5b
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B |
| chr15_-_76084035 | 0.20 |
ENSMUST00000054449.14
ENSMUST00000169714.8 ENSMUST00000165453.8 |
Plec
|
plectin |
| chr9_+_109760931 | 0.20 |
ENSMUST00000165876.8
|
Map4
|
microtubule-associated protein 4 |
| chr16_-_85347305 | 0.20 |
ENSMUST00000175700.8
ENSMUST00000114174.3 |
Cyyr1
|
cysteine and tyrosine-rich protein 1 |
| chr19_-_7318798 | 0.20 |
ENSMUST00000165965.8
ENSMUST00000051711.16 ENSMUST00000169541.8 ENSMUST00000165989.2 |
Mark2
|
MAP/microtubule affinity regulating kinase 2 |
| chr14_+_45457168 | 0.20 |
ENSMUST00000227086.2
ENSMUST00000147957.2 |
Gpr137c
|
G protein-coupled receptor 137C |
| chr2_+_156263002 | 0.20 |
ENSMUST00000125153.10
ENSMUST00000103136.8 ENSMUST00000109577.9 |
Epb41l1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr2_+_68691902 | 0.20 |
ENSMUST00000176018.2
|
Cers6
|
ceramide synthase 6 |
| chr1_+_60137412 | 0.20 |
ENSMUST00000124986.8
|
Carf
|
calcium response factor |
| chr11_+_77873535 | 0.20 |
ENSMUST00000108360.8
ENSMUST00000049167.14 |
Phf12
|
PHD finger protein 12 |
| chr11_+_49094292 | 0.20 |
ENSMUST00000150284.8
ENSMUST00000109197.8 ENSMUST00000151228.2 |
Zfp62
|
zinc finger protein 62 |
| chr18_-_4352944 | 0.20 |
ENSMUST00000025078.10
|
Map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr17_+_47451868 | 0.20 |
ENSMUST00000190080.9
|
Trerf1
|
transcriptional regulating factor 1 |
| chr14_+_25459690 | 0.20 |
ENSMUST00000007961.15
|
Zmiz1
|
zinc finger, MIZ-type containing 1 |
| chr15_-_102165884 | 0.20 |
ENSMUST00000043172.15
|
Rarg
|
retinoic acid receptor, gamma |
| chr8_+_4288815 | 0.20 |
ENSMUST00000003027.14
ENSMUST00000110999.8 |
Map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr11_+_24028173 | 0.19 |
ENSMUST00000109514.8
|
Bcl11a
|
B cell CLL/lymphoma 11A (zinc finger protein) |
| chr8_-_85526972 | 0.19 |
ENSMUST00000099070.10
|
Nfix
|
nuclear factor I/X |
| chr7_-_103320398 | 0.19 |
ENSMUST00000062144.4
|
Olfr624
|
olfactory receptor 624 |
| chr19_-_17814984 | 0.19 |
ENSMUST00000025618.16
ENSMUST00000050715.10 |
Pcsk5
|
proprotein convertase subtilisin/kexin type 5 |
| chr16_-_59421342 | 0.19 |
ENSMUST00000172910.3
|
Crybg3
|
beta-gamma crystallin domain containing 3 |
| chr14_+_27150750 | 0.19 |
ENSMUST00000022450.6
|
Tasor
|
transcription activation suppressor |
| chr5_+_123280250 | 0.19 |
ENSMUST00000174836.8
ENSMUST00000163030.9 |
Setd1b
|
SET domain containing 1B |
| chr9_+_109760528 | 0.19 |
ENSMUST00000035055.15
|
Map4
|
microtubule-associated protein 4 |
| chr17_-_66079629 | 0.19 |
ENSMUST00000233258.2
|
Rab31
|
RAB31, member RAS oncogene family |
| chrX_-_20816841 | 0.19 |
ENSMUST00000009550.14
|
Elk1
|
ELK1, member of ETS oncogene family |
| chr15_-_50752437 | 0.19 |
ENSMUST00000183997.8
ENSMUST00000183757.8 |
Trps1
|
transcriptional repressor GATA binding 1 |
| chr5_+_32293145 | 0.18 |
ENSMUST00000031017.11
|
Fosl2
|
fos-like antigen 2 |
| chr6_-_48422759 | 0.18 |
ENSMUST00000114561.9
|
Zfp467
|
zinc finger protein 467 |
| chr6_-_48422307 | 0.18 |
ENSMUST00000114563.8
ENSMUST00000114558.8 ENSMUST00000101443.10 |
Zfp467
|
zinc finger protein 467 |
| chr5_-_102217770 | 0.18 |
ENSMUST00000053177.14
ENSMUST00000174698.2 |
Wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr8_-_106198112 | 0.18 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr14_+_76652369 | 0.18 |
ENSMUST00000110888.8
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr15_+_78784043 | 0.18 |
ENSMUST00000001226.11
|
Sh3bp1
|
SH3-domain binding protein 1 |
| chrX_+_105070865 | 0.18 |
ENSMUST00000113557.8
|
Atp7a
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr15_-_103242697 | 0.18 |
ENSMUST00000229373.2
|
Zfp385a
|
zinc finger protein 385A |
| chr18_-_80756273 | 0.18 |
ENSMUST00000078049.12
ENSMUST00000236711.2 |
Nfatc1
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1 |
| chr17_-_85995680 | 0.17 |
ENSMUST00000024947.8
ENSMUST00000163568.4 |
Six2
|
sine oculis-related homeobox 2 |
| chr16_+_24212284 | 0.17 |
ENSMUST00000038053.14
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chrX_+_162692126 | 0.17 |
ENSMUST00000033734.14
ENSMUST00000112294.9 |
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chrX_+_7523499 | 0.17 |
ENSMUST00000033485.14
|
Prickle3
|
prickle planar cell polarity protein 3 |
| chrX_+_105070907 | 0.17 |
ENSMUST00000055941.7
|
Atp7a
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr18_+_42644552 | 0.17 |
ENSMUST00000237602.2
ENSMUST00000236088.2 ENSMUST00000025375.15 |
Tcerg1
|
transcription elongation regulator 1 (CA150) |
| chr6_-_39183712 | 0.17 |
ENSMUST00000002305.9
|
Kdm7a
|
lysine (K)-specific demethylase 7A |
| chr17_+_35455532 | 0.17 |
ENSMUST00000068261.9
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2 |
| chr7_+_24048613 | 0.17 |
ENSMUST00000032683.6
|
Lypd5
|
Ly6/Plaur domain containing 5 |
| chr11_+_24028022 | 0.17 |
ENSMUST00000000881.13
|
Bcl11a
|
B cell CLL/lymphoma 11A (zinc finger protein) |
| chr13_+_31809774 | 0.16 |
ENSMUST00000042054.3
|
Foxf2
|
forkhead box F2 |
| chr19_-_7319157 | 0.16 |
ENSMUST00000164205.8
ENSMUST00000165286.8 ENSMUST00000168324.2 ENSMUST00000032557.15 |
Mark2
|
MAP/microtubule affinity regulating kinase 2 |
| chr15_+_30172716 | 0.16 |
ENSMUST00000081728.7
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2 |
| chr19_+_23736205 | 0.16 |
ENSMUST00000025830.9
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr2_-_32631028 | 0.16 |
ENSMUST00000009695.3
|
6330409D20Rik
|
RIKEN cDNA 6330409D20 gene |
| chr7_+_100355798 | 0.16 |
ENSMUST00000107042.9
ENSMUST00000207564.2 ENSMUST00000049053.9 |
Fam168a
|
family with sequence similarity 168, member A |
| chr7_-_48530777 | 0.16 |
ENSMUST00000058745.15
|
E2f8
|
E2F transcription factor 8 |
| chr11_-_50844572 | 0.16 |
ENSMUST00000162420.2
ENSMUST00000051159.3 |
Prop1
|
paired like homeodomain factor 1 |
| chr5_+_29639662 | 0.16 |
ENSMUST00000001611.11
|
Nom1
|
nucleolar protein with MIF4G domain 1 |
| chr6_+_4601124 | 0.16 |
ENSMUST00000141359.2
|
Casd1
|
CAS1 domain containing 1 |
| chr9_-_43017249 | 0.16 |
ENSMUST00000165665.9
|
Arhgef12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr14_+_25459630 | 0.16 |
ENSMUST00000162645.8
|
Zmiz1
|
zinc finger, MIZ-type containing 1 |
| chr8_-_85526653 | 0.16 |
ENSMUST00000126806.2
ENSMUST00000076715.13 |
Nfix
|
nuclear factor I/X |
| chr6_-_38276157 | 0.16 |
ENSMUST00000058524.3
|
Zc3hav1l
|
zinc finger CCCH-type, antiviral 1-like |
| chr4_-_126096112 | 0.16 |
ENSMUST00000142125.2
ENSMUST00000106141.3 |
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr19_+_4203603 | 0.15 |
ENSMUST00000236632.2
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C polypeptide-associated protein |
| chr17_-_24428351 | 0.15 |
ENSMUST00000024931.6
|
Ntn3
|
netrin 3 |
| chr17_+_31120785 | 0.15 |
ENSMUST00000114574.3
|
Glp1r
|
glucagon-like peptide 1 receptor |
| chr1_-_64776890 | 0.15 |
ENSMUST00000116133.4
ENSMUST00000063982.7 |
Fzd5
|
frizzled class receptor 5 |
| chr3_-_89959739 | 0.15 |
ENSMUST00000199929.2
ENSMUST00000090908.11 ENSMUST00000198322.5 ENSMUST00000196843.5 |
Ubap2l
|
ubiquitin-associated protein 2-like |
| chr6_-_48422612 | 0.15 |
ENSMUST00000114556.2
|
Zfp467
|
zinc finger protein 467 |
| chr7_-_48531344 | 0.15 |
ENSMUST00000119223.2
|
E2f8
|
E2F transcription factor 8 |
| chr5_+_135197137 | 0.15 |
ENSMUST00000031692.12
|
Bcl7b
|
B cell CLL/lymphoma 7B |
| chr7_-_92319096 | 0.15 |
ENSMUST00000208255.2
ENSMUST00000208058.2 |
Pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr1_-_74002156 | 0.15 |
ENSMUST00000191367.2
|
Tns1
|
tensin 1 |
| chr11_+_74721733 | 0.15 |
ENSMUST00000000291.9
|
Mnt
|
max binding protein |
| chr11_+_49094119 | 0.15 |
ENSMUST00000109198.8
ENSMUST00000137061.9 |
Zfp62
|
zinc finger protein 62 |
| chr1_-_72576089 | 0.15 |
ENSMUST00000047786.6
|
Marchf4
|
membrane associated ring-CH-type finger 4 |
| chr17_+_5045178 | 0.15 |
ENSMUST00000092723.11
ENSMUST00000232180.2 ENSMUST00000115797.9 |
Arid1b
|
AT rich interactive domain 1B (SWI-like) |
| chr2_+_69477552 | 0.15 |
ENSMUST00000074963.9
ENSMUST00000112286.9 |
Bbs5
|
Bardet-Biedl syndrome 5 (human) |
| chr18_+_82929451 | 0.15 |
ENSMUST00000235902.2
|
Zfp516
|
zinc finger protein 516 |
| chr1_-_173707677 | 0.15 |
ENSMUST00000190651.4
ENSMUST00000188804.7 |
Mndal
|
myeloid nuclear differentiation antigen like |
| chr15_+_12117899 | 0.14 |
ENSMUST00000122941.8
|
Zfr
|
zinc finger RNA binding protein |
| chr5_-_138270995 | 0.14 |
ENSMUST00000161665.2
ENSMUST00000100530.8 ENSMUST00000161279.8 ENSMUST00000161647.8 |
Gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr7_+_24982206 | 0.14 |
ENSMUST00000165239.3
|
Cic
|
capicua transcriptional repressor |
| chr11_+_112673041 | 0.14 |
ENSMUST00000000579.3
|
Sox9
|
SRY (sex determining region Y)-box 9 |
| chr1_-_155912159 | 0.14 |
ENSMUST00000097527.10
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr7_-_4400704 | 0.14 |
ENSMUST00000108590.4
ENSMUST00000206928.2 |
Gp6
|
glycoprotein 6 (platelet) |
| chr9_-_57743989 | 0.14 |
ENSMUST00000164010.8
ENSMUST00000171444.8 ENSMUST00000098686.4 |
Arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr5_-_90031180 | 0.14 |
ENSMUST00000198151.2
|
Adamts3
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 3 |
| chr9_-_56325344 | 0.14 |
ENSMUST00000061552.15
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr11_+_74816640 | 0.14 |
ENSMUST00000045281.13
|
Smg6
|
Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) |
| chr1_-_173770010 | 0.14 |
ENSMUST00000042228.15
ENSMUST00000081216.12 ENSMUST00000129829.8 ENSMUST00000123708.8 |
Ifi203
|
interferon activated gene 203 |
| chr14_+_21550921 | 0.14 |
ENSMUST00000182964.3
|
Kat6b
|
K(lysine) acetyltransferase 6B |
| chr9_+_27702243 | 0.14 |
ENSMUST00000115243.9
|
Opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr1_-_155848917 | 0.14 |
ENSMUST00000138762.8
|
Cep350
|
centrosomal protein 350 |
| chr16_-_76169902 | 0.14 |
ENSMUST00000054178.8
|
Nrip1
|
nuclear receptor interacting protein 1 |
| chr15_-_102154874 | 0.14 |
ENSMUST00000063339.14
|
Rarg
|
retinoic acid receptor, gamma |
| chr9_-_107749594 | 0.14 |
ENSMUST00000183035.2
|
Rbm6
|
RNA binding motif protein 6 |
| chr19_+_6413840 | 0.13 |
ENSMUST00000113488.8
ENSMUST00000113487.8 |
Sf1
|
splicing factor 1 |
| chr9_+_72182142 | 0.13 |
ENSMUST00000184036.8
ENSMUST00000184517.8 ENSMUST00000098576.10 |
Zfp280d
|
zinc finger protein 280D |
| chr2_+_158509039 | 0.13 |
ENSMUST00000045503.11
|
Ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr19_-_27988393 | 0.13 |
ENSMUST00000172907.8
ENSMUST00000046898.17 |
Rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr8_+_4288733 | 0.13 |
ENSMUST00000110998.9
ENSMUST00000062686.11 |
Map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr19_-_59931432 | 0.13 |
ENSMUST00000170819.2
|
Rab11fip2
|
RAB11 family interacting protein 2 (class I) |
| chr8_-_85500998 | 0.13 |
ENSMUST00000109762.8
|
Nfix
|
nuclear factor I/X |
| chr11_+_78006391 | 0.13 |
ENSMUST00000155571.2
|
Fam222b
|
family with sequence similarity 222, member B |
| chr10_+_59715378 | 0.13 |
ENSMUST00000147914.8
ENSMUST00000146590.8 |
Dnajb12
|
DnaJ heat shock protein family (Hsp40) member B12 |
| chr3_-_89959770 | 0.13 |
ENSMUST00000029553.16
ENSMUST00000195995.5 ENSMUST00000064639.15 ENSMUST00000199834.5 |
Ubap2l
|
ubiquitin-associated protein 2-like |
| chr17_+_34811217 | 0.13 |
ENSMUST00000038149.13
|
Pbx2
|
pre B cell leukemia homeobox 2 |
| chr6_+_37507108 | 0.13 |
ENSMUST00000040987.11
|
Akr1d1
|
aldo-keto reductase family 1, member D1 |
| chrX_+_167819606 | 0.13 |
ENSMUST00000087016.11
ENSMUST00000112129.8 ENSMUST00000112131.9 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr6_+_124986224 | 0.13 |
ENSMUST00000112427.8
|
Zfp384
|
zinc finger protein 384 |
| chr13_-_96678844 | 0.13 |
ENSMUST00000223475.2
|
Polk
|
polymerase (DNA directed), kappa |
| chr3_-_89294430 | 0.13 |
ENSMUST00000107433.8
|
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr7_-_92319126 | 0.13 |
ENSMUST00000119954.9
|
Pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr5_-_107437427 | 0.13 |
ENSMUST00000031224.15
|
Tgfbr3
|
transforming growth factor, beta receptor III |
| chr14_+_30673334 | 0.13 |
ENSMUST00000226551.2
ENSMUST00000228328.2 |
Nek4
|
NIMA (never in mitosis gene a)-related expressed kinase 4 |
| chr7_+_28869629 | 0.12 |
ENSMUST00000098609.4
|
Ggn
|
gametogenetin |
| chr5_-_115479203 | 0.12 |
ENSMUST00000139167.3
|
Gatc
|
glutamyl-tRNA(Gln) amidotransferase, subunit C |
| chr7_-_28135616 | 0.12 |
ENSMUST00000208199.2
|
Samd4b
|
sterile alpha motif domain containing 4B |
| chr11_+_84771133 | 0.12 |
ENSMUST00000020837.7
|
Myo19
|
myosin XIX |
| chr7_+_92524495 | 0.12 |
ENSMUST00000207594.2
|
Prcp
|
prolylcarboxypeptidase (angiotensinase C) |
| chr9_+_107217786 | 0.12 |
ENSMUST00000042581.4
|
6430571L13Rik
|
RIKEN cDNA 6430571L13 gene |
| chr4_+_125384481 | 0.12 |
ENSMUST00000030676.8
|
Grik3
|
glutamate receptor, ionotropic, kainate 3 |
| chr1_+_23801007 | 0.12 |
ENSMUST00000063663.6
|
B3gat2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr2_-_37312881 | 0.12 |
ENSMUST00000112936.4
ENSMUST00000112934.8 |
Rc3h2
|
ring finger and CCCH-type zinc finger domains 2 |
| chr7_+_28988724 | 0.12 |
ENSMUST00000207714.2
ENSMUST00000048187.6 |
Ppp1r14a
|
protein phosphatase 1, regulatory inhibitor subunit 14A |
| chr19_+_6413703 | 0.12 |
ENSMUST00000131252.8
ENSMUST00000113489.8 |
Sf1
|
splicing factor 1 |
| chr15_-_78456898 | 0.12 |
ENSMUST00000043214.8
|
Rac2
|
Rac family small GTPase 2 |
| chr11_-_98040377 | 0.12 |
ENSMUST00000103143.10
|
Fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr16_+_80997580 | 0.12 |
ENSMUST00000037785.14
ENSMUST00000067602.5 |
Ncam2
|
neural cell adhesion molecule 2 |
| chr6_+_124986078 | 0.12 |
ENSMUST00000054553.11
|
Zfp384
|
zinc finger protein 384 |
| chr5_-_137739364 | 0.12 |
ENSMUST00000149512.3
|
Nyap1
|
neuronal tyrosine-phosphorylated phosphoinositide 3-kinase adaptor 1 |
| chr14_+_121272950 | 0.12 |
ENSMUST00000026635.8
|
Farp1
|
FERM, RhoGEF (Arhgef) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr17_+_24768808 | 0.12 |
ENSMUST00000228550.2
ENSMUST00000035565.5 |
Pkd1
|
polycystin 1, transient receptor poteintial channel interacting |
| chr19_-_29782907 | 0.12 |
ENSMUST00000175726.8
|
9930021J03Rik
|
RIKEN cDNA 9930021J03 gene |
| chr7_-_97387429 | 0.12 |
ENSMUST00000206389.2
|
Aqp11
|
aquaporin 11 |
| chr9_+_72182248 | 0.12 |
ENSMUST00000183410.8
|
Zfp280d
|
zinc finger protein 280D |
| chr18_+_35695736 | 0.12 |
ENSMUST00000235851.2
ENSMUST00000235581.2 |
Matr3
|
matrin 3 |
| chr15_+_102178975 | 0.12 |
ENSMUST00000181801.2
|
Gm9918
|
predicted gene 9918 |
| chr17_+_81251997 | 0.12 |
ENSMUST00000025092.5
|
Tmem178
|
transmembrane protein 178 |
| chr7_+_101619327 | 0.11 |
ENSMUST00000210475.2
|
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr8_+_47192767 | 0.11 |
ENSMUST00000034041.9
ENSMUST00000208507.2 ENSMUST00000207105.2 |
Irf2
|
interferon regulatory factor 2 |
| chr17_-_45046499 | 0.11 |
ENSMUST00000162373.8
ENSMUST00000162878.8 |
Runx2
|
runt related transcription factor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp263
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.2 | 0.6 | GO:0070949 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.1 | 0.6 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.1 | 0.4 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.1 | 0.6 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.1 | 0.4 | GO:1904959 | elastin biosynthetic process(GO:0051542) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.1 | 0.5 | GO:1904799 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) |
| 0.1 | 0.3 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.1 | 0.3 | GO:0072076 | hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme development(GO:0072076) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.1 | 0.3 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.1 | 0.6 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.1 | 0.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.3 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 0.3 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.5 | GO:1900019 | prolactin secretion(GO:0070459) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.1 | 0.2 | GO:0002353 | plasma kallikrein-kinin cascade(GO:0002353) |
| 0.1 | 0.3 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.1 | 0.3 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 0.2 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 0.4 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 0.2 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
| 0.1 | 0.2 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.1 | 0.2 | GO:0043380 | regulation of memory T cell differentiation(GO:0043380) |
| 0.1 | 0.2 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.1 | 0.2 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 0.2 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.2 | GO:0060061 | Spemann organizer formation(GO:0060061) syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.0 | 0.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.2 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.2 | GO:0032887 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.2 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 0.0 | 0.1 | GO:1900062 | regulation of replicative cell aging(GO:1900062) |
| 0.0 | 0.1 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.2 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.0 | 0.1 | GO:0060939 | transforming growth factor beta receptor complex assembly(GO:0007181) cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.1 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.0 | 0.1 | GO:1900126 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.0 | 0.1 | GO:0061289 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.0 | 0.0 | GO:0043585 | nose morphogenesis(GO:0043585) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.0 | 0.3 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.1 | GO:0071317 | cellular response to isoquinoline alkaloid(GO:0071317) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.4 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:1990164 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.2 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.1 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.0 | 0.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.3 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.3 | GO:1901250 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.0 | 0.3 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) regulation of rRNA processing(GO:2000232) |
| 0.0 | 0.1 | GO:0042376 | menaquinone metabolic process(GO:0009233) phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.1 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.5 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.0 | 0.4 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.2 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.0 | 0.2 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.1 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.0 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.2 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.0 | 0.2 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.0 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.0 | 0.4 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.0 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.3 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 0.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.1 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 0.2 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.3 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.0 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.3 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.0 | GO:1904828 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.0 | GO:0021998 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.0 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.3 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.2 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.0 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.0 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.8 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.0 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.4 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.8 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.2 | GO:0097635 | Atg12-Atg5-Atg16 complex(GO:0034274) extrinsic component of autophagosome membrane(GO:0097635) |
| 0.1 | 0.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.3 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.4 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.0 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.3 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.1 | 0.4 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.1 | 0.3 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 0.2 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.1 | 1.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.3 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.6 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.2 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.3 | GO:0044547 | rRNA primary transcript binding(GO:0042134) DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.2 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0035671 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) |
| 0.0 | 0.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.2 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.0 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0004985 | melanocortin receptor activity(GO:0004977) opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0038064 | collagen receptor activity(GO:0038064) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.6 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.5 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |