Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
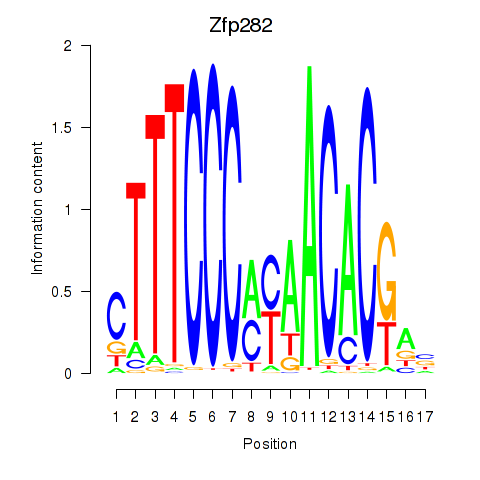
Results for Zfp282
Z-value: 0.99
Transcription factors associated with Zfp282
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp282
|
ENSMUSG00000025821.10 | zinc finger protein 282 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp282 | mm39_v1_chr6_+_47854138_47854138 | 0.23 | 7.0e-01 | Click! |
Activity profile of Zfp282 motif
Sorted Z-values of Zfp282 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_35255729 | 0.74 |
ENSMUST00000040962.6
|
Nudt19
|
nudix (nucleoside diphosphate linked moiety X)-type motif 19 |
| chr4_+_41760454 | 0.64 |
ENSMUST00000108040.8
|
Il11ra1
|
interleukin 11 receptor, alpha chain 1 |
| chr12_+_87490666 | 0.58 |
ENSMUST00000161023.8
ENSMUST00000160488.8 ENSMUST00000077462.8 ENSMUST00000160880.2 |
Slirp
|
SRA stem-loop interacting RNA binding protein |
| chr16_+_10884156 | 0.54 |
ENSMUST00000089011.6
|
Snn
|
stannin |
| chr2_-_91274967 | 0.53 |
ENSMUST00000064652.14
ENSMUST00000102594.11 ENSMUST00000094835.9 |
1110051M20Rik
|
RIKEN cDNA 1110051M20 gene |
| chr1_-_125362321 | 0.52 |
ENSMUST00000191544.7
|
Actr3
|
ARP3 actin-related protein 3 |
| chr4_+_102843540 | 0.51 |
ENSMUST00000030248.12
ENSMUST00000125417.9 ENSMUST00000169211.3 |
Dynlt5
|
dynein light chain Tctex-type 5 |
| chr11_-_68864666 | 0.50 |
ENSMUST00000038644.5
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr1_+_134383247 | 0.50 |
ENSMUST00000112232.8
ENSMUST00000027725.11 ENSMUST00000116528.2 |
Klhl12
|
kelch-like 12 |
| chr12_+_71021395 | 0.43 |
ENSMUST00000160027.8
ENSMUST00000160864.8 |
Psma3
|
proteasome subunit alpha 3 |
| chr7_+_142558783 | 0.43 |
ENSMUST00000009396.13
|
Tspan32
|
tetraspanin 32 |
| chr3_-_88858465 | 0.43 |
ENSMUST00000173135.8
|
Dap3
|
death associated protein 3 |
| chr8_-_23727639 | 0.42 |
ENSMUST00000033950.7
|
Gins4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr7_-_143294051 | 0.40 |
ENSMUST00000119499.8
|
Osbpl5
|
oxysterol binding protein-like 5 |
| chr11_-_120687195 | 0.39 |
ENSMUST00000143139.8
ENSMUST00000129955.2 ENSMUST00000026151.11 ENSMUST00000167023.8 ENSMUST00000106133.8 ENSMUST00000106135.8 |
Dus1l
|
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr19_-_21418215 | 0.38 |
ENSMUST00000237823.2
|
Gda
|
guanine deaminase |
| chr7_+_142558837 | 0.38 |
ENSMUST00000207211.2
|
Tspan32
|
tetraspanin 32 |
| chr1_+_36800874 | 0.37 |
ENSMUST00000027291.7
|
Zap70
|
zeta-chain (TCR) associated protein kinase |
| chr14_+_119175328 | 0.33 |
ENSMUST00000022734.9
|
Dnajc3
|
DnaJ heat shock protein family (Hsp40) member C3 |
| chr3_-_88858402 | 0.33 |
ENSMUST00000173021.8
|
Dap3
|
death associated protein 3 |
| chr1_+_151220222 | 0.32 |
ENSMUST00000023918.13
ENSMUST00000111887.10 ENSMUST00000097543.8 |
Ivns1abp
|
influenza virus NS1A binding protein |
| chr2_+_35512023 | 0.31 |
ENSMUST00000091010.12
|
Dab2ip
|
disabled 2 interacting protein |
| chr9_-_114610879 | 0.30 |
ENSMUST00000084867.9
ENSMUST00000216760.2 ENSMUST00000035009.16 |
Cmtm7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr10_+_93289264 | 0.29 |
ENSMUST00000016033.9
|
Lta4h
|
leukotriene A4 hydrolase |
| chr7_+_101732323 | 0.27 |
ENSMUST00000124189.3
|
Trpc2
|
transient receptor potential cation channel, subfamily C, member 2 |
| chr9_-_105008972 | 0.27 |
ENSMUST00000186925.2
|
Nudt16
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16 |
| chr2_+_35512172 | 0.26 |
ENSMUST00000112992.9
|
Dab2ip
|
disabled 2 interacting protein |
| chr1_+_52884172 | 0.25 |
ENSMUST00000159352.8
ENSMUST00000044478.7 |
Hibch
|
3-hydroxyisobutyryl-Coenzyme A hydrolase |
| chr13_-_9815173 | 0.25 |
ENSMUST00000062658.15
ENSMUST00000222358.2 |
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr4_-_40239778 | 0.24 |
ENSMUST00000037907.13
|
Ddx58
|
DEAD/H box helicase 58 |
| chr13_-_9815350 | 0.23 |
ENSMUST00000110636.9
ENSMUST00000152725.8 |
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr19_-_6835538 | 0.22 |
ENSMUST00000113440.2
|
Ccdc88b
|
coiled-coil domain containing 88B |
| chr7_-_90106375 | 0.22 |
ENSMUST00000032844.7
|
Tmem126a
|
transmembrane protein 126A |
| chr5_-_65548723 | 0.19 |
ENSMUST00000120094.5
ENSMUST00000127874.6 ENSMUST00000057885.13 |
Rpl9
|
ribosomal protein L9 |
| chr10_+_20024203 | 0.19 |
ENSMUST00000020173.16
|
Map7
|
microtubule-associated protein 7 |
| chr9_-_105009023 | 0.19 |
ENSMUST00000035179.9
|
Nudt16
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16 |
| chr13_-_9815276 | 0.18 |
ENSMUST00000130151.8
|
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr8_+_3726503 | 0.17 |
ENSMUST00000207787.2
|
Trappc5
|
trafficking protein particle complex 5 |
| chr3_+_154302311 | 0.17 |
ENSMUST00000192462.6
ENSMUST00000029850.15 |
Cryz
|
crystallin, zeta |
| chr12_-_87490580 | 0.17 |
ENSMUST00000162961.8
ENSMUST00000185301.2 |
Alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr7_+_16472335 | 0.17 |
ENSMUST00000086112.8
ENSMUST00000205607.2 |
Ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr5_+_65548840 | 0.17 |
ENSMUST00000122026.6
ENSMUST00000031101.10 ENSMUST00000200374.2 |
Lias
|
lipoic acid synthetase |
| chr3_-_88857707 | 0.17 |
ENSMUST00000090938.11
|
Dap3
|
death associated protein 3 |
| chr5_-_122270014 | 0.16 |
ENSMUST00000128101.8
ENSMUST00000132701.2 |
Ccdc63
|
coiled-coil domain containing 63 |
| chr16_+_87251852 | 0.16 |
ENSMUST00000119504.8
ENSMUST00000131356.8 |
Usp16
|
ubiquitin specific peptidase 16 |
| chrY_-_90850446 | 0.16 |
ENSMUST00000179623.2
|
Gm21748
|
predicted gene, 21748 |
| chr12_+_72583114 | 0.15 |
ENSMUST00000044352.8
|
Pcnx4
|
pecanex homolog 4 |
| chr12_+_111504450 | 0.15 |
ENSMUST00000166123.9
ENSMUST00000222441.2 |
Eif5
|
eukaryotic translation initiation factor 5 |
| chr11_-_102298748 | 0.14 |
ENSMUST00000124755.2
|
Slc25a39
|
solute carrier family 25, member 39 |
| chr3_+_93349637 | 0.14 |
ENSMUST00000064257.6
|
Tchh
|
trichohyalin |
| chr7_-_105289515 | 0.14 |
ENSMUST00000133519.8
ENSMUST00000209550.2 ENSMUST00000210911.2 ENSMUST00000084782.10 ENSMUST00000131446.8 |
Arfip2
|
ADP-ribosylation factor interacting protein 2 |
| chr9_-_83028523 | 0.14 |
ENSMUST00000187193.7
ENSMUST00000185315.7 |
Hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr2_+_24276545 | 0.13 |
ENSMUST00000127242.2
|
Psd4
|
pleckstrin and Sec7 domain containing 4 |
| chr4_+_14864076 | 0.13 |
ENSMUST00000029875.4
|
Pip4p2
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 2 |
| chr13_-_53531391 | 0.13 |
ENSMUST00000021920.8
|
Sptlc1
|
serine palmitoyltransferase, long chain base subunit 1 |
| chr7_+_105289729 | 0.13 |
ENSMUST00000210350.2
ENSMUST00000151193.2 ENSMUST00000209588.2 ENSMUST00000106780.2 ENSMUST00000106784.2 ENSMUST00000106785.8 ENSMUST00000106786.8 ENSMUST00000211054.2 |
Timm10b
Gm45799
|
translocase of inner mitochondrial membrane 10B predicted gene 45799 |
| chr5_-_129907878 | 0.11 |
ENSMUST00000026617.13
|
Phkg1
|
phosphorylase kinase gamma 1 |
| chr7_-_45175570 | 0.10 |
ENSMUST00000167273.2
ENSMUST00000042105.11 |
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr17_-_33904345 | 0.10 |
ENSMUST00000234474.2
ENSMUST00000139302.8 ENSMUST00000114385.9 |
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr9_+_108782664 | 0.10 |
ENSMUST00000026740.6
|
Col7a1
|
collagen, type VII, alpha 1 |
| chr17_-_50401305 | 0.10 |
ENSMUST00000113195.8
|
Rftn1
|
raftlin lipid raft linker 1 |
| chr7_-_10229249 | 0.10 |
ENSMUST00000032551.8
|
Zik1
|
zinc finger protein interacting with K protein 1 |
| chr11_-_102120917 | 0.10 |
ENSMUST00000008999.12
|
Hdac5
|
histone deacetylase 5 |
| chr19_-_18609118 | 0.10 |
ENSMUST00000025631.7
ENSMUST00000236615.2 |
Ostf1
|
osteoclast stimulating factor 1 |
| chr11_-_102120953 | 0.09 |
ENSMUST00000107150.8
ENSMUST00000156337.2 ENSMUST00000107151.9 ENSMUST00000107152.9 |
Hdac5
|
histone deacetylase 5 |
| chr6_-_39787813 | 0.09 |
ENSMUST00000114797.2
ENSMUST00000031978.9 |
Mrps33
|
mitochondrial ribosomal protein S33 |
| chr9_-_85631361 | 0.09 |
ENSMUST00000039213.15
|
Ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr12_+_111504640 | 0.08 |
ENSMUST00000222375.2
ENSMUST00000222388.2 |
Eif5
|
eukaryotic translation initiation factor 5 |
| chr17_-_33904386 | 0.08 |
ENSMUST00000087582.13
|
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr19_-_5538370 | 0.07 |
ENSMUST00000124334.8
|
Mus81
|
MUS81 structure-specific endonuclease subunit |
| chr10_+_20024724 | 0.07 |
ENSMUST00000116259.5
ENSMUST00000214231.2 |
Map7
|
microtubule-associated protein 7 |
| chr7_+_107264518 | 0.06 |
ENSMUST00000239294.2
|
Ppfibp2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr8_+_26298502 | 0.05 |
ENSMUST00000033979.6
|
Star
|
steroidogenic acute regulatory protein |
| chr11_+_9068098 | 0.05 |
ENSMUST00000020677.8
ENSMUST00000101525.9 |
Upp1
|
uridine phosphorylase 1 |
| chr2_+_3771709 | 0.05 |
ENSMUST00000177037.2
|
Fam107b
|
family with sequence similarity 107, member B |
| chr11_-_109188892 | 0.05 |
ENSMUST00000106706.8
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr4_-_148244028 | 0.04 |
ENSMUST00000167160.8
ENSMUST00000151246.8 |
Fbxo44
|
F-box protein 44 |
| chr13_+_23343482 | 0.04 |
ENSMUST00000226845.2
ENSMUST00000228666.2 ENSMUST00000227388.2 |
Vmn1r219
|
vomeronasal 1 receptor 219 |
| chr14_+_31058589 | 0.04 |
ENSMUST00000022451.14
|
Capn7
|
calpain 7 |
| chr15_-_103337982 | 0.04 |
ENSMUST00000146736.8
|
Gtsf1
|
gametocyte specific factor 1 |
| chr17_+_46991926 | 0.04 |
ENSMUST00000233491.2
|
Mea1
|
male enhanced antigen 1 |
| chr10_-_19727165 | 0.03 |
ENSMUST00000059805.6
|
Slc35d3
|
solute carrier family 35, member D3 |
| chr13_-_22289994 | 0.03 |
ENSMUST00000227357.2
ENSMUST00000228428.2 |
Vmn1r189
|
vomeronasal 1 receptor 189 |
| chr11_+_9068507 | 0.03 |
ENSMUST00000164791.8
ENSMUST00000130522.8 |
Upp1
|
uridine phosphorylase 1 |
| chr9_+_38738911 | 0.03 |
ENSMUST00000051238.7
ENSMUST00000219798.2 |
Olfr923
|
olfactory receptor 923 |
| chr7_+_105289655 | 0.02 |
ENSMUST00000058333.10
|
Timm10b
|
translocase of inner mitochondrial membrane 10B |
| chr7_-_29772226 | 0.02 |
ENSMUST00000183115.8
ENSMUST00000182919.8 ENSMUST00000183190.2 ENSMUST00000080834.15 |
Zfp82
|
zinc finger protein 82 |
| chr6_+_68518603 | 0.02 |
ENSMUST00000168090.3
ENSMUST00000103326.3 |
Igkv1-99
|
immunoglobulin kappa variable 1-99 |
| chrX_+_134894573 | 0.02 |
ENSMUST00000058119.9
|
Arxes2
|
adipocyte-related X-chromosome expressed sequence 2 |
| chr7_-_47849036 | 0.02 |
ENSMUST00000094388.3
|
Mrgprb4
|
MAS-related GPR, member B4 |
| chr18_-_35855383 | 0.02 |
ENSMUST00000133064.8
|
Ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr11_+_9068134 | 0.02 |
ENSMUST00000170444.8
|
Upp1
|
uridine phosphorylase 1 |
| chr2_-_125466985 | 0.02 |
ENSMUST00000089776.3
|
Cep152
|
centrosomal protein 152 |
| chr5_-_87638728 | 0.02 |
ENSMUST00000147854.6
|
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr14_+_66207163 | 0.02 |
ENSMUST00000153460.8
|
Clu
|
clusterin |
| chr3_-_108306267 | 0.02 |
ENSMUST00000147251.2
|
Celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr6_-_128503666 | 0.01 |
ENSMUST00000143664.2
ENSMUST00000112132.8 |
Pzp
|
PZP, alpha-2-macroglobulin like |
| chr17_+_46991350 | 0.01 |
ENSMUST00000232732.2
|
Mea1
|
male enhanced antigen 1 |
| chr2_-_88818544 | 0.01 |
ENSMUST00000099804.2
|
Olfr1214
|
olfactory receptor 1214 |
| chr19_+_13140364 | 0.01 |
ENSMUST00000207113.3
|
Olfr1461
|
olfactory receptor 1461 |
| chr17_+_38378064 | 0.01 |
ENSMUST00000087129.3
|
Olfr130
|
olfactory receptor 130 |
| chr9_+_45341589 | 0.01 |
ENSMUST00000239471.2
ENSMUST00000034592.11 ENSMUST00000239429.2 |
Dscaml1
|
DS cell adhesion molecule like 1 |
| chr18_-_78222349 | 0.01 |
ENSMUST00000237290.2
|
Slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr16_+_7011580 | 0.01 |
ENSMUST00000231194.2
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr1_+_87522267 | 0.01 |
ENSMUST00000165109.2
ENSMUST00000070898.6 |
Neu2
|
neuraminidase 2 |
| chr7_-_106291021 | 0.01 |
ENSMUST00000216118.3
|
Olfr694
|
olfactory receptor 694 |
| chr4_-_148244489 | 0.01 |
ENSMUST00000057907.10
|
Fbxo44
|
F-box protein 44 |
| chr8_-_93774820 | 0.00 |
ENSMUST00000095211.5
|
Ces1a
|
carboxylesterase 1A |
| chr16_-_85599994 | 0.00 |
ENSMUST00000023610.15
|
Adamts1
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 1 |
| chr19_+_5385672 | 0.00 |
ENSMUST00000043380.5
|
Catsper1
|
cation channel, sperm associated 1 |
| chrY_-_2170393 | 0.00 |
ENSMUST00000115891.2
|
Zfy2
|
zinc finger protein 2, Y-linked |
| chr4_-_88562696 | 0.00 |
ENSMUST00000105149.3
|
Ifna13
|
interferon alpha 13 |
| chr3_-_92577621 | 0.00 |
ENSMUST00000029530.6
|
Lce1a2
|
late cornified envelope 1A2 |
| chr6_-_83504756 | 0.00 |
ENSMUST00000152029.2
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr2_-_160754212 | 0.00 |
ENSMUST00000109454.8
ENSMUST00000057169.5 |
Emilin3
|
elastin microfibril interfacer 3 |
| chrX_+_104543374 | 0.00 |
ENSMUST00000041758.5
|
Cypt2
|
cysteine-rich perinuclear theca 2 |
| chr7_+_26456567 | 0.00 |
ENSMUST00000077855.8
|
Cyp2b19
|
cytochrome P450, family 2, subfamily b, polypeptide 19 |
| chrX_+_102458066 | 0.00 |
ENSMUST00000033691.5
|
Tsx
|
testis specific X-linked gene |
| chr11_+_49170977 | 0.00 |
ENSMUST00000078932.2
|
Olfr1393
|
olfactory receptor 1393 |
| chr14_+_41000762 | 0.00 |
ENSMUST00000172412.2
|
Gm2832
|
predicted gene 2832 |
| chr14_+_75479727 | 0.00 |
ENSMUST00000022576.10
|
Cpb2
|
carboxypeptidase B2 (plasma) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp282
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.6 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 | 0.5 | GO:1904171 | negative regulation of bleb assembly(GO:1904171) |
| 0.1 | 0.4 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.1 | 0.5 | GO:0046707 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.1 | 0.5 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.3 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.8 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.1 | 0.3 | GO:0036493 | positive regulation of translational initiation in response to stress(GO:0032058) positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.1 | 0.4 | GO:0043366 | beta selection(GO:0043366) |
| 0.1 | 0.2 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.1 | 0.5 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.1 | 0.5 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.3 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.2 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) RNA repair(GO:0042245) |
| 0.0 | 0.2 | GO:0070537 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.1 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.0 | 0.1 | GO:0046108 | uridine catabolic process(GO:0006218) uridine metabolic process(GO:0046108) |
| 0.0 | 0.4 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.3 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.7 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.3 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.2 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 0.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.0 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.2 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0070442 | integrin alphaIIb-beta3 complex(GO:0070442) |
| 0.1 | 0.6 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.1 | 0.4 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.3 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.9 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.2 | 0.5 | GO:0035870 | dITP diphosphatase activity(GO:0035870) |
| 0.1 | 0.4 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.5 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.3 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.2 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.0 | 0.2 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.2 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.2 | GO:0043734 | DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.0 | 0.5 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.1 | GO:0048256 | flap endonuclease activity(GO:0048256) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |