Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
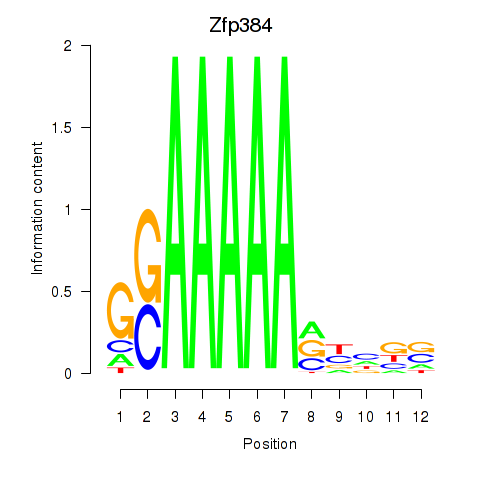
Results for Zfp384
Z-value: 1.82
Transcription factors associated with Zfp384
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp384
|
ENSMUSG00000038346.19 | zinc finger protein 384 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp384 | mm39_v1_chr6_+_124986193_124986216 | -0.58 | 3.0e-01 | Click! |
Activity profile of Zfp384 motif
Sorted Z-values of Zfp384 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_21934675 | 1.60 |
ENSMUST00000102983.2
|
H4c12
|
H4 clustered histone 12 |
| chr13_-_23806530 | 1.34 |
ENSMUST00000062045.4
|
H1f4
|
H1.4 linker histone, cluster member |
| chr6_+_88175312 | 1.24 |
ENSMUST00000203480.2
ENSMUST00000015197.9 |
Gata2
|
GATA binding protein 2 |
| chr3_-_20296337 | 1.09 |
ENSMUST00000001921.3
|
Cpa3
|
carboxypeptidase A3, mast cell |
| chr13_+_23759930 | 1.08 |
ENSMUST00000105105.4
|
H3c4
|
H3 clustered histone 4 |
| chr13_+_23728222 | 1.06 |
ENSMUST00000075558.5
|
H3c7
|
H3 clustered histone 7 |
| chr13_+_23922783 | 1.02 |
ENSMUST00000040914.3
|
H1f2
|
H1.2 linker histone, cluster member |
| chr13_-_23758370 | 0.87 |
ENSMUST00000105106.2
|
H2bc7
|
H2B clustered histone 7 |
| chr13_+_23718038 | 0.85 |
ENSMUST00000073261.3
|
H2ac10
|
H2A clustered histone 10 |
| chr6_+_129326927 | 0.78 |
ENSMUST00000065289.6
|
Clec12a
|
C-type lectin domain family 12, member a |
| chr13_+_21971631 | 0.74 |
ENSMUST00000110473.3
ENSMUST00000102982.2 |
H2bc22
|
H2B clustered histone 22 |
| chr13_-_23929490 | 0.71 |
ENSMUST00000091752.5
|
H3c3
|
H3 clustered histone 3 |
| chr13_+_23715220 | 0.63 |
ENSMUST00000102972.6
|
H4c8
|
H4 clustered histone 8 |
| chr13_+_23947641 | 0.58 |
ENSMUST00000055770.4
|
H1f1
|
H1.1 linker histone, cluster member |
| chr6_+_146697539 | 0.56 |
ENSMUST00000111639.8
|
Arntl2
|
aryl hydrocarbon receptor nuclear translocator-like 2 |
| chr11_-_43317017 | 0.56 |
ENSMUST00000101340.11
ENSMUST00000118368.8 ENSMUST00000020685.10 ENSMUST00000020687.15 |
Pttg1
|
pituitary tumor-transforming gene 1 |
| chr10_+_128736827 | 0.55 |
ENSMUST00000219317.2
|
Cd63
|
CD63 antigen |
| chr13_-_23727549 | 0.52 |
ENSMUST00000224359.2
|
H2bc9
|
H2B clustered histone 9 |
| chr13_-_21964747 | 0.48 |
ENSMUST00000080511.3
|
H1f5
|
H1.5 linker histone, cluster member |
| chr7_-_45159790 | 0.47 |
ENSMUST00000211765.2
|
Nucb1
|
nucleobindin 1 |
| chr15_-_74983786 | 0.46 |
ENSMUST00000191451.2
ENSMUST00000100542.10 |
Ly6c2
|
lymphocyte antigen 6 complex, locus C2 |
| chr7_-_19426529 | 0.46 |
ENSMUST00000207978.2
ENSMUST00000108451.4 ENSMUST00000045035.12 |
Apoc1
|
apolipoprotein C-I |
| chr11_+_54793569 | 0.45 |
ENSMUST00000082430.11
|
Gpx3
|
glutathione peroxidase 3 |
| chr14_+_12016304 | 0.45 |
ENSMUST00000161302.8
|
Fhit
|
fragile histidine triad gene |
| chr3_+_151143557 | 0.45 |
ENSMUST00000196970.3
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr13_-_21971388 | 0.44 |
ENSMUST00000091751.3
|
H2ac22
|
H2A clustered histone 22 |
| chrX_+_93768175 | 0.41 |
ENSMUST00000101388.4
|
Zxdb
|
zinc finger, X-linked, duplicated B |
| chr17_-_50401305 | 0.41 |
ENSMUST00000113195.8
|
Rftn1
|
raftlin lipid raft linker 1 |
| chr3_+_5815863 | 0.41 |
ENSMUST00000192045.2
|
Gm8797
|
predicted pseudogene 8797 |
| chr10_-_111833138 | 0.40 |
ENSMUST00000074805.12
|
Glipr1
|
GLI pathogenesis-related 1 (glioma) |
| chr7_-_3551003 | 0.39 |
ENSMUST00000065703.9
ENSMUST00000203020.3 ENSMUST00000203821.3 |
Tarm1
|
T cell-interacting, activating receptor on myeloid cells 1 |
| chr13_+_23758555 | 0.38 |
ENSMUST00000090776.7
|
H2ac7
|
H2A clustered histone 7 |
| chr6_+_135339929 | 0.38 |
ENSMUST00000032330.16
|
Emp1
|
epithelial membrane protein 1 |
| chr19_-_40260286 | 0.38 |
ENSMUST00000182432.2
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr1_+_88015524 | 0.35 |
ENSMUST00000113139.2
|
Ugt1a8
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr17_+_43978377 | 0.33 |
ENSMUST00000233627.2
ENSMUST00000233437.2 |
Cyp39a1
|
cytochrome P450, family 39, subfamily a, polypeptide 1 |
| chr13_+_23935088 | 0.31 |
ENSMUST00000078369.3
|
H2ac4
|
H2A clustered histone 4 |
| chr6_-_52181393 | 0.30 |
ENSMUST00000048794.7
|
Hoxa5
|
homeobox A5 |
| chr15_+_95698574 | 0.30 |
ENSMUST00000226793.2
|
Ano6
|
anoctamin 6 |
| chr19_-_40260060 | 0.30 |
ENSMUST00000068439.13
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr9_-_54467419 | 0.30 |
ENSMUST00000041901.7
|
Cib2
|
calcium and integrin binding family member 2 |
| chr7_-_19410749 | 0.30 |
ENSMUST00000003074.16
|
Apoc2
|
apolipoprotein C-II |
| chr17_+_43978280 | 0.29 |
ENSMUST00000170988.2
|
Cyp39a1
|
cytochrome P450, family 39, subfamily a, polypeptide 1 |
| chr9_+_118892497 | 0.28 |
ENSMUST00000141185.8
ENSMUST00000126251.8 ENSMUST00000136561.2 |
Vill
|
villin-like |
| chrM_+_3906 | 0.28 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr13_-_35211060 | 0.28 |
ENSMUST00000170538.8
ENSMUST00000163280.8 |
Eci2
|
enoyl-Coenzyme A delta isomerase 2 |
| chr5_+_16758538 | 0.28 |
ENSMUST00000199581.5
|
Hgf
|
hepatocyte growth factor |
| chr10_-_128245501 | 0.28 |
ENSMUST00000172348.8
ENSMUST00000166608.8 ENSMUST00000164199.8 ENSMUST00000171370.2 ENSMUST00000026439.14 |
Nabp2
|
nucleic acid binding protein 2 |
| chr11_+_54988866 | 0.27 |
ENSMUST00000000608.8
|
Gm2a
|
GM2 ganglioside activator protein |
| chr1_+_194621127 | 0.27 |
ENSMUST00000016638.8
ENSMUST00000110815.9 |
Cd34
|
CD34 antigen |
| chr4_-_131565542 | 0.27 |
ENSMUST00000030741.9
ENSMUST00000105987.9 |
Ptpru
|
protein tyrosine phosphatase, receptor type, U |
| chr2_+_164782642 | 0.27 |
ENSMUST00000137626.2
|
Mmp9
|
matrix metallopeptidase 9 |
| chr1_-_128520002 | 0.26 |
ENSMUST00000052172.7
ENSMUST00000142893.2 |
Cxcr4
|
chemokine (C-X-C motif) receptor 4 |
| chr16_+_35977170 | 0.26 |
ENSMUST00000079184.6
|
Stfa2l1
|
stefin A2 like 1 |
| chr3_-_15397325 | 0.26 |
ENSMUST00000108361.2
|
Gm9733
|
predicted gene 9733 |
| chr19_-_33567708 | 0.26 |
ENSMUST00000112508.9
|
Lipo3
|
lipase, member O3 |
| chr15_-_102140331 | 0.26 |
ENSMUST00000230652.2
ENSMUST00000127014.3 ENSMUST00000001327.11 |
Itgb7
|
integrin beta 7 |
| chr6_+_124489364 | 0.25 |
ENSMUST00000068593.9
|
C1ra
|
complement component 1, r subcomponent A |
| chrX_+_106192510 | 0.25 |
ENSMUST00000147521.8
ENSMUST00000167673.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr10_-_81335966 | 0.25 |
ENSMUST00000053646.7
|
S1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr8_+_95721378 | 0.25 |
ENSMUST00000212956.2
ENSMUST00000212531.2 |
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr9_+_14411907 | 0.24 |
ENSMUST00000004200.9
|
Cwc15
|
CWC15 spliceosome-associated protein |
| chr1_+_156193607 | 0.24 |
ENSMUST00000102782.4
|
Gm2000
|
predicted gene 2000 |
| chr17_+_31329939 | 0.24 |
ENSMUST00000236241.2
|
Abcg1
|
ATP binding cassette subfamily G member 1 |
| chr15_-_74855264 | 0.24 |
ENSMUST00000023250.11
ENSMUST00000166694.2 |
Ly6i
|
lymphocyte antigen 6 complex, locus I |
| chr9_+_14411956 | 0.24 |
ENSMUST00000215143.2
|
Cwc15
|
CWC15 spliceosome-associated protein |
| chr11_-_69586626 | 0.24 |
ENSMUST00000108649.3
ENSMUST00000174159.8 ENSMUST00000181810.8 |
Tnfsfm13
Tnfsf12
|
tumor necrosis factor (ligand) superfamily, membrane-bound member 13 tumor necrosis factor (ligand) superfamily, member 12 |
| chr15_+_102234589 | 0.24 |
ENSMUST00000170627.8
|
Pfdn5
|
prefoldin 5 |
| chr13_-_25015392 | 0.24 |
ENSMUST00000006900.7
|
Acot13
|
acyl-CoA thioesterase 13 |
| chr3_-_107925122 | 0.23 |
ENSMUST00000126593.3
|
Gstm1
|
glutathione S-transferase, mu 1 |
| chr13_-_22219738 | 0.23 |
ENSMUST00000091742.6
|
H2ac12
|
H2A clustered histone 12 |
| chrX_-_52672363 | 0.23 |
ENSMUST00000088778.5
|
Rtl8b
|
retrotransposon Gag like 8B |
| chr11_+_62442502 | 0.23 |
ENSMUST00000136938.2
|
Ubb
|
ubiquitin B |
| chr13_-_49369454 | 0.23 |
ENSMUST00000048946.7
|
Card19
|
caspase recruitment domain family, member 19 |
| chr11_+_62737936 | 0.23 |
ENSMUST00000150989.8
ENSMUST00000176577.2 |
Fbxw10
|
F-box and WD-40 domain protein 10 |
| chr14_+_25842565 | 0.23 |
ENSMUST00000022416.15
|
Anxa11
|
annexin A11 |
| chrX_+_150784841 | 0.23 |
ENSMUST00000026289.10
ENSMUST00000112617.4 |
Hsd17b10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr1_-_171061902 | 0.23 |
ENSMUST00000079957.12
|
Fcer1g
|
Fc receptor, IgE, high affinity I, gamma polypeptide |
| chr2_-_126460575 | 0.23 |
ENSMUST00000028838.5
|
Hdc
|
histidine decarboxylase |
| chr7_+_4340708 | 0.22 |
ENSMUST00000006792.6
ENSMUST00000126417.3 |
Ncr1
|
natural cytotoxicity triggering receptor 1 |
| chr6_+_48578935 | 0.22 |
ENSMUST00000204095.3
|
Zfp775
|
zinc finger protein 775 |
| chr5_+_135698881 | 0.22 |
ENSMUST00000153500.8
|
Por
|
P450 (cytochrome) oxidoreductase |
| chr3_+_89173859 | 0.22 |
ENSMUST00000040824.2
|
Dpm3
|
dolichyl-phosphate mannosyltransferase polypeptide 3 |
| chr11_+_119804630 | 0.22 |
ENSMUST00000026434.13
ENSMUST00000124199.8 |
Chmp6
|
charged multivesicular body protein 6 |
| chr14_+_24540777 | 0.22 |
ENSMUST00000169826.3
ENSMUST00000225023.2 ENSMUST00000223999.2 |
Rps24
|
ribosomal protein S24 |
| chr16_+_93404719 | 0.22 |
ENSMUST00000039659.9
ENSMUST00000231762.2 |
Cbr1
|
carbonyl reductase 1 |
| chr1_+_172327569 | 0.22 |
ENSMUST00000111230.8
|
Tagln2
|
transgelin 2 |
| chr8_-_84299540 | 0.22 |
ENSMUST00000212990.2
|
Tecr
|
trans-2,3-enoyl-CoA reductase |
| chr12_-_85421467 | 0.21 |
ENSMUST00000040766.9
|
Tmed10
|
transmembrane p24 trafficking protein 10 |
| chr11_-_62680273 | 0.21 |
ENSMUST00000054654.13
|
Zfp286
|
zinc finger protein 286 |
| chr10_-_127016448 | 0.21 |
ENSMUST00000222911.3
ENSMUST00000095270.3 |
Slc26a10
|
solute carrier family 26, member 10 |
| chr13_+_23879775 | 0.21 |
ENSMUST00000041052.5
|
H1f6
|
H1.6 linker histone, cluster member |
| chr7_+_45683122 | 0.21 |
ENSMUST00000033121.7
|
Nomo1
|
nodal modulator 1 |
| chr1_+_134487893 | 0.21 |
ENSMUST00000047714.14
|
Kdm5b
|
lysine (K)-specific demethylase 5B |
| chr11_+_70416185 | 0.21 |
ENSMUST00000018430.7
|
Psmb6
|
proteasome (prosome, macropain) subunit, beta type 6 |
| chr16_-_36228798 | 0.21 |
ENSMUST00000023619.8
|
Stfa2
|
stefin A2 |
| chr15_+_102234551 | 0.21 |
ENSMUST00000166658.8
ENSMUST00000062492.11 |
Pfdn5
|
prefoldin 5 |
| chr16_-_36188086 | 0.21 |
ENSMUST00000096089.3
|
Cstdc5
|
cystatin domain containing 5 |
| chr6_-_134609350 | 0.21 |
ENSMUST00000047443.5
|
Mansc1
|
MANSC domain containing 1 |
| chr4_-_57301791 | 0.20 |
ENSMUST00000075637.11
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr6_+_138117519 | 0.20 |
ENSMUST00000120939.8
ENSMUST00000204628.3 ENSMUST00000140932.2 ENSMUST00000120302.8 |
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr17_+_56056977 | 0.20 |
ENSMUST00000025004.7
|
Adgre4
|
adhesion G protein-coupled receptor E4 |
| chr6_-_38814159 | 0.20 |
ENSMUST00000160962.8
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr6_-_115785695 | 0.20 |
ENSMUST00000081840.6
|
Rpl32
|
ribosomal protein L32 |
| chr5_-_121731615 | 0.20 |
ENSMUST00000129753.8
ENSMUST00000152945.3 |
Aldh2
|
aldehyde dehydrogenase 2, mitochondrial |
| chr7_-_28465870 | 0.20 |
ENSMUST00000085851.12
ENSMUST00000032815.11 |
Nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, beta |
| chr16_+_33650002 | 0.20 |
ENSMUST00000115028.11
ENSMUST00000069345.6 |
Itgb5
|
integrin beta 5 |
| chr15_+_102234825 | 0.20 |
ENSMUST00000165671.8
ENSMUST00000165717.4 |
Pfdn5
|
prefoldin 5 |
| chr2_-_21210151 | 0.20 |
ENSMUST00000027992.3
|
Enkur
|
enkurin, TRPC channel interacting protein |
| chr3_+_7568481 | 0.20 |
ENSMUST00000051064.9
ENSMUST00000193010.2 |
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr16_+_17615146 | 0.20 |
ENSMUST00000012161.5
ENSMUST00000232577.2 |
Scarf2
|
scavenger receptor class F, member 2 |
| chr13_+_100806206 | 0.20 |
ENSMUST00000170347.4
ENSMUST00000057325.15 |
Ccdc125
|
coiled-coil domain containing 125 |
| chr17_+_18108086 | 0.20 |
ENSMUST00000149944.2
|
Fpr2
|
formyl peptide receptor 2 |
| chr11_-_115382622 | 0.20 |
ENSMUST00000106530.8
ENSMUST00000021082.7 |
Nt5c
|
5',3'-nucleotidase, cytosolic |
| chr15_+_95688712 | 0.20 |
ENSMUST00000071874.8
|
Ano6
|
anoctamin 6 |
| chr7_-_29898236 | 0.20 |
ENSMUST00000001845.13
|
Capns1
|
calpain, small subunit 1 |
| chr17_-_26727437 | 0.19 |
ENSMUST00000236661.2
ENSMUST00000025025.7 |
Dusp1
|
dual specificity phosphatase 1 |
| chr17_+_47083561 | 0.19 |
ENSMUST00000071430.7
|
2310039H08Rik
|
RIKEN cDNA 2310039H08 gene |
| chr7_+_126295114 | 0.19 |
ENSMUST00000106369.2
|
Bola2
|
bolA-like 2 (E. coli) |
| chr4_+_156077834 | 0.19 |
ENSMUST00000105578.2
|
Sdf4
|
stromal cell derived factor 4 |
| chr4_+_123798690 | 0.19 |
ENSMUST00000106202.4
|
Mycbp
|
MYC binding protein |
| chr9_+_89936581 | 0.19 |
ENSMUST00000143172.8
ENSMUST00000185459.2 |
Ctsh
|
cathepsin H |
| chr8_-_106718334 | 0.19 |
ENSMUST00000117555.8
ENSMUST00000034373.8 |
Dpep2
|
dipeptidase 2 |
| chr5_-_104169696 | 0.19 |
ENSMUST00000119025.2
|
Hsd17b11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr1_-_84262274 | 0.18 |
ENSMUST00000177458.2
ENSMUST00000168574.9 |
Pid1
|
phosphotyrosine interaction domain containing 1 |
| chr8_+_95703728 | 0.18 |
ENSMUST00000179619.9
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr8_-_70963202 | 0.18 |
ENSMUST00000125184.8
|
Uba52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr11_-_78875689 | 0.18 |
ENSMUST00000108269.10
ENSMUST00000108268.10 |
Lgals9
|
lectin, galactose binding, soluble 9 |
| chr11_-_100628979 | 0.18 |
ENSMUST00000155500.2
ENSMUST00000107364.8 ENSMUST00000019317.12 |
Rab5c
|
RAB5C, member RAS oncogene family |
| chr12_+_108145802 | 0.18 |
ENSMUST00000221167.2
|
Ccnk
|
cyclin K |
| chr5_+_135093056 | 0.18 |
ENSMUST00000071263.7
|
Dnajc30
|
DnaJ heat shock protein family (Hsp40) member C30 |
| chr11_-_78074377 | 0.18 |
ENSMUST00000102483.5
|
Rpl23a
|
ribosomal protein L23A |
| chr1_-_171910324 | 0.18 |
ENSMUST00000003550.11
|
Ncstn
|
nicastrin |
| chr11_+_54793743 | 0.18 |
ENSMUST00000149324.3
|
Gpx3
|
glutathione peroxidase 3 |
| chr9_+_14411921 | 0.18 |
ENSMUST00000213913.2
|
Cwc15
|
CWC15 spliceosome-associated protein |
| chr7_+_30021165 | 0.18 |
ENSMUST00000098585.4
|
E130208F15Rik
|
RIKEN cDNA E130208F15 gene |
| chr6_+_39358036 | 0.18 |
ENSMUST00000031986.5
|
Rab19
|
RAB19, member RAS oncogene family |
| chr14_-_73622638 | 0.18 |
ENSMUST00000228637.2
ENSMUST00000022704.9 |
Itm2b
|
integral membrane protein 2B |
| chr16_+_33071784 | 0.17 |
ENSMUST00000023502.6
|
Snx4
|
sorting nexin 4 |
| chr7_+_28466160 | 0.17 |
ENSMUST00000122915.8
ENSMUST00000072965.5 ENSMUST00000170068.9 |
Sirt2
|
sirtuin 2 |
| chr14_-_31807552 | 0.17 |
ENSMUST00000022461.11
ENSMUST00000067955.12 ENSMUST00000124303.9 |
Dph3
|
diphthamine biosynthesis 3 |
| chr2_+_120398168 | 0.17 |
ENSMUST00000028743.10
ENSMUST00000116437.8 ENSMUST00000153580.8 ENSMUST00000142278.2 |
Snap23
|
synaptosomal-associated protein 23 |
| chr4_+_138032035 | 0.17 |
ENSMUST00000030538.5
|
Ddost
|
dolichyl-di-phosphooligosaccharide-protein glycotransferase |
| chr6_-_52195663 | 0.17 |
ENSMUST00000134367.4
|
Hoxa7
|
homeobox A7 |
| chr11_+_62737887 | 0.17 |
ENSMUST00000036085.11
|
Fbxw10
|
F-box and WD-40 domain protein 10 |
| chr14_-_70666513 | 0.17 |
ENSMUST00000226426.2
ENSMUST00000048129.6 |
Piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr4_+_102835888 | 0.17 |
ENSMUST00000223169.2
|
Dynlt5
|
dynein light chain Tctex-type 5 |
| chr7_+_126294527 | 0.17 |
ENSMUST00000130498.2
|
Bola2
|
bolA-like 2 (E. coli) |
| chr12_+_108145997 | 0.17 |
ENSMUST00000101055.5
|
Ccnk
|
cyclin K |
| chr15_-_101262452 | 0.17 |
ENSMUST00000230909.2
|
Krt80
|
keratin 80 |
| chr8_+_26210064 | 0.17 |
ENSMUST00000068916.16
ENSMUST00000139836.8 |
Plpp5
|
phospholipid phosphatase 5 |
| chr12_+_112645237 | 0.17 |
ENSMUST00000174780.2
ENSMUST00000169593.2 ENSMUST00000173942.2 |
Zbtb42
|
zinc finger and BTB domain containing 42 |
| chrX_+_93679671 | 0.17 |
ENSMUST00000096368.4
|
Gspt2
|
G1 to S phase transition 2 |
| chr1_+_74275648 | 0.17 |
ENSMUST00000113820.9
ENSMUST00000006467.14 |
Arpc2
|
actin related protein 2/3 complex, subunit 2 |
| chr2_+_4923815 | 0.17 |
ENSMUST00000027975.8
|
Phyh
|
phytanoyl-CoA hydroxylase |
| chr7_+_43751749 | 0.17 |
ENSMUST00000085455.6
|
Klk1b21
|
kallikrein 1-related peptidase b21 |
| chr2_-_127050161 | 0.17 |
ENSMUST00000056146.2
|
1810024B03Rik
|
RIKEN cDNA 1810024B03 gene |
| chr7_-_126165530 | 0.17 |
ENSMUST00000180459.3
ENSMUST00000032992.7 |
Eif3c
|
eukaryotic translation initiation factor 3, subunit C |
| chr4_-_44073016 | 0.17 |
ENSMUST00000128439.8
ENSMUST00000140724.3 |
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr9_+_40597297 | 0.17 |
ENSMUST00000034522.8
|
Clmp
|
CXADR-like membrane protein |
| chr16_+_32876813 | 0.17 |
ENSMUST00000078804.12
ENSMUST00000115079.8 |
Rpl35a
|
ribosomal protein L35A |
| chr1_-_88437617 | 0.17 |
ENSMUST00000067625.9
|
Glrp1
|
glutamine repeat protein 1 |
| chr10_+_128583734 | 0.16 |
ENSMUST00000163377.10
|
Pym1
|
PYM homolog 1, exon junction complex associated factor |
| chr13_+_49340961 | 0.16 |
ENSMUST00000049022.15
|
Ninj1
|
ninjurin 1 |
| chr9_-_20726344 | 0.16 |
ENSMUST00000004201.8
|
Col5a3
|
collagen, type V, alpha 3 |
| chr18_+_9707595 | 0.16 |
ENSMUST00000234965.2
|
Colec12
|
collectin sub-family member 12 |
| chr13_+_9326513 | 0.16 |
ENSMUST00000174552.8
|
Dip2c
|
disco interacting protein 2 homolog C |
| chr9_+_115738523 | 0.16 |
ENSMUST00000119291.8
ENSMUST00000069651.13 |
Gadl1
|
glutamate decarboxylase-like 1 |
| chr4_-_148021217 | 0.16 |
ENSMUST00000019199.14
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr11_-_46203047 | 0.16 |
ENSMUST00000129474.2
ENSMUST00000093166.11 ENSMUST00000165599.9 |
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr7_+_16186704 | 0.16 |
ENSMUST00000019302.10
|
Tmem160
|
transmembrane protein 160 |
| chr11_-_94283792 | 0.16 |
ENSMUST00000178136.8
ENSMUST00000021231.8 |
Abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr8_+_12997948 | 0.16 |
ENSMUST00000110871.8
|
Mcf2l
|
mcf.2 transforming sequence-like |
| chr19_-_24938909 | 0.16 |
ENSMUST00000025815.10
|
Cbwd1
|
COBW domain containing 1 |
| chr17_-_79292856 | 0.16 |
ENSMUST00000118991.2
|
Prkd3
|
protein kinase D3 |
| chr16_+_36004452 | 0.16 |
ENSMUST00000114858.2
|
Cstdc4
|
cystatin domain containing 4 |
| chr8_-_45786226 | 0.16 |
ENSMUST00000095328.6
|
Cyp4v3
|
cytochrome P450, family 4, subfamily v, polypeptide 3 |
| chr14_-_14371476 | 0.15 |
ENSMUST00000225653.2
ENSMUST00000112689.9 ENSMUST00000036682.9 |
Pxk
|
PX domain containing serine/threonine kinase |
| chr1_-_153425791 | 0.15 |
ENSMUST00000041874.9
|
Npl
|
N-acetylneuraminate pyruvate lyase |
| chr16_-_15681211 | 0.15 |
ENSMUST00000023351.11
ENSMUST00000117136.2 |
Mzt2
|
mitotic spindle organizing protein 2 |
| chr5_-_129856237 | 0.15 |
ENSMUST00000118268.9
|
Psph
|
phosphoserine phosphatase |
| chr8_-_71219299 | 0.15 |
ENSMUST00000222087.4
|
Ifi30
|
interferon gamma inducible protein 30 |
| chr1_-_69723316 | 0.15 |
ENSMUST00000190855.7
ENSMUST00000188110.7 ENSMUST00000191262.7 |
Ikzf2
|
IKAROS family zinc finger 2 |
| chr19_-_44017637 | 0.15 |
ENSMUST00000026211.10
ENSMUST00000211830.2 |
Cyp2c23
|
cytochrome P450, family 2, subfamily c, polypeptide 23 |
| chr11_+_69805005 | 0.15 |
ENSMUST00000057884.6
|
Gps2
|
G protein pathway suppressor 2 |
| chr2_+_90865958 | 0.15 |
ENSMUST00000111445.10
ENSMUST00000111446.10 ENSMUST00000050323.6 |
Rapsn
|
receptor-associated protein of the synapse |
| chrX_+_106193060 | 0.15 |
ENSMUST00000125676.8
ENSMUST00000180182.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr1_-_150341911 | 0.15 |
ENSMUST00000162367.8
ENSMUST00000161611.8 ENSMUST00000161320.8 ENSMUST00000159035.2 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr14_-_47655621 | 0.15 |
ENSMUST00000180299.8
|
Dlgap5
|
DLG associated protein 5 |
| chr7_-_46569617 | 0.15 |
ENSMUST00000210664.2
ENSMUST00000156335.9 |
Tsg101
|
tumor susceptibility gene 101 |
| chrX_-_69520778 | 0.15 |
ENSMUST00000101506.10
ENSMUST00000114630.3 |
Tmem185a
|
transmembrane protein 185A |
| chr16_+_38383182 | 0.15 |
ENSMUST00000163948.8
|
Tmem39a
|
transmembrane protein 39a |
| chr11_+_62441992 | 0.15 |
ENSMUST00000019649.4
|
Ubb
|
ubiquitin B |
| chr18_+_75138910 | 0.15 |
ENSMUST00000040284.6
ENSMUST00000236421.2 ENSMUST00000237263.2 |
BC031181
|
cDNA sequence BC031181 |
| chr3_-_137687284 | 0.15 |
ENSMUST00000136613.4
ENSMUST00000029806.13 |
Dapp1
|
dual adaptor for phosphotyrosine and 3-phosphoinositides 1 |
| chr19_+_12824046 | 0.15 |
ENSMUST00000189517.2
|
Gm5244
|
predicted pseudogene 5244 |
| chr19_+_53131187 | 0.15 |
ENSMUST00000050096.15
ENSMUST00000237832.2 |
Add3
|
adducin 3 (gamma) |
| chr7_+_127661835 | 0.15 |
ENSMUST00000106242.10
ENSMUST00000120355.8 ENSMUST00000106240.9 ENSMUST00000098015.10 |
Itgam
Gm49368
|
integrin alpha M predicted gene, 49368 |
| chr13_-_119545479 | 0.15 |
ENSMUST00000223268.2
|
Nnt
|
nicotinamide nucleotide transhydrogenase |
| chr5_-_125467240 | 0.15 |
ENSMUST00000156249.2
|
Ubc
|
ubiquitin C |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp384
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0035854 | eosinophil fate commitment(GO:0035854) |
| 0.2 | 0.6 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.2 | 0.2 | GO:0010813 | neuropeptide catabolic process(GO:0010813) |
| 0.2 | 0.5 | GO:0097045 | activation of blood coagulation via clotting cascade(GO:0002543) phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.1 | 0.4 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.1 | 0.2 | GO:0035938 | estradiol secretion(GO:0035938) regulation of estradiol secretion(GO:2000864) |
| 0.1 | 0.3 | GO:0060435 | bronchiole development(GO:0060435) intestinal epithelial cell maturation(GO:0060574) |
| 0.1 | 1.1 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.1 | 0.3 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.5 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 0.3 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 0.3 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.1 | 0.3 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.1 | 0.3 | GO:0036301 | macrophage colony-stimulating factor production(GO:0036301) granulocyte colony-stimulating factor production(GO:0071611) regulation of granulocyte colony-stimulating factor production(GO:0071655) regulation of macrophage colony-stimulating factor production(GO:1901256) |
| 0.1 | 0.2 | GO:0016132 | brassinosteroid metabolic process(GO:0016131) brassinosteroid biosynthetic process(GO:0016132) |
| 0.1 | 0.1 | GO:2001189 | negative regulation of immunological synapse formation(GO:2000521) regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001188) negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001189) |
| 0.1 | 0.3 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.1 | 0.4 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.1 | 0.6 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.1 | 0.2 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.1 | 0.2 | GO:0009223 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.1 | 0.2 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.1 | 0.2 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.1 | 0.4 | GO:0002554 | serotonin secretion by platelet(GO:0002554) |
| 0.1 | 0.2 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.1 | 0.3 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.2 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.1 | 0.3 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.1 | 0.2 | GO:0043602 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.1 | 0.2 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.1 | 0.2 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.1 | 0.2 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 0.4 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.3 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.0 | 0.2 | GO:0070625 | zymogen granule exocytosis(GO:0070625) |
| 0.0 | 0.2 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.4 | GO:0002441 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.1 | GO:1904266 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.0 | 0.2 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.0 | 0.4 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 0.4 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.4 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.2 | GO:0000239 | pachytene(GO:0000239) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.1 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.0 | 0.2 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.1 | GO:0007225 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.0 | 0.5 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.2 | GO:0034436 | glycoprotein transport(GO:0034436) response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.3 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0090673 | endothelial cell-matrix adhesion(GO:0090673) |
| 0.0 | 0.1 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.0 | 0.3 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.3 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.2 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:1903094 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.2 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.0 | 0.1 | GO:0070194 | synaptonemal complex disassembly(GO:0070194) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.7 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.1 | GO:0052564 | response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.0 | 0.1 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) |
| 0.0 | 0.1 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.1 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0071877 | regulation of adrenergic receptor signaling pathway(GO:0071877) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.1 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.0 | 0.3 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0097325 | regulation of collagen catabolic process(GO:0010710) melanocyte proliferation(GO:0097325) |
| 0.0 | 0.0 | GO:0051794 | regulation of catagen(GO:0051794) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.2 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.7 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.1 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.1 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.3 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.2 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.1 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.1 | GO:0071640 | regulation of macrophage inflammatory protein 1 alpha production(GO:0071640) |
| 0.0 | 0.1 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.1 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0019427 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.1 | GO:2000328 | positive regulation of memory T cell differentiation(GO:0043382) regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.0 | 0.1 | GO:1902477 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.0 | 0.2 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.1 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.0 | 0.1 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:1903121 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) |
| 0.0 | 0.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.3 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.0 | 0.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.2 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.3 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.7 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0048208 | vesicle coating(GO:0006901) vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.0 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.0 | GO:0070340 | detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.1 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.1 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.1 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.3 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.0 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.0 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.0 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.1 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.2 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.0 | 0.1 | GO:0097298 | regulation of nucleus size(GO:0097298) |
| 0.0 | 0.2 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.2 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.0 | 0.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:1902109 | granzyme-mediated apoptotic signaling pathway(GO:0008626) negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:0098961 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.0 | 0.1 | GO:1900004 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.3 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.0 | 0.2 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 | 0.3 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.0 | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 0.0 | 0.1 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.3 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0060931 | sinoatrial node cell differentiation(GO:0060921) sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.0 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.1 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.0 | 0.0 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.0 | GO:0036258 | multivesicular body assembly(GO:0036258) |
| 0.0 | 0.1 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.0 | GO:0002838 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) |
| 0.0 | 0.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.0 | GO:0050717 | positive regulation of interleukin-1 alpha production(GO:0032730) positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.0 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.0 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.1 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.0 | GO:0046967 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) cytosol to ER transport(GO:0046967) |
| 0.0 | 0.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.0 | GO:0090157 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) modulation by virus of host autophagy(GO:0039519) suppression by virus of host autophagy(GO:0039521) amino acid homeostasis(GO:0080144) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) |
| 0.0 | 0.0 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.0 | GO:0051466 | positive regulation of corticotropin-releasing hormone secretion(GO:0051466) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.1 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 0.4 | GO:0002945 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.1 | 0.2 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.1 | 0.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.6 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.5 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.7 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0044317 | rod spherule(GO:0044317) |
| 0.0 | 0.5 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.2 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 0.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.5 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.2 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.6 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 1.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.8 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.0 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.6 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.0 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.1 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.0 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.0 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0008396 | oxysterol 7-alpha-hydroxylase activity(GO:0008396) |
| 0.1 | 0.6 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.1 | 0.4 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.4 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.1 | 0.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.4 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.1 | 0.3 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.1 | 0.2 | GO:0047598 | sterol delta7 reductase activity(GO:0009918) 7-dehydrocholesterol reductase activity(GO:0047598) |
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.2 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.1 | 0.2 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.1 | 0.2 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.1 | 1.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 0.3 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 0.2 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.1 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.3 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.1 | 0.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.2 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 0.0 | 0.2 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.5 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 1.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.6 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.6 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.2 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.3 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.2 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.3 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.1 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.4 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0071796 | K6-linked polyubiquitin binding(GO:0071796) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.0 | 0.2 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.0 | 0.1 | GO:0015205 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.6 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.1 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.1 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.0 | 0.3 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.0 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.0 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.0 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.3 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.0 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.0 | 0.0 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.1 | GO:0051870 | methotrexate binding(GO:0051870) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 2.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 0.8 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.1 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.5 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.1 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 1.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |