Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
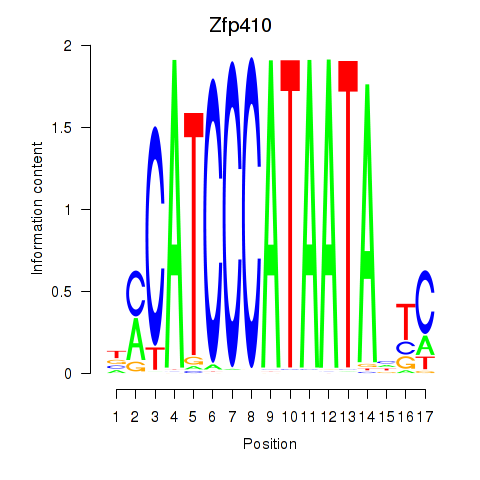
Results for Zfp410
Z-value: 0.41
Transcription factors associated with Zfp410
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp410
|
ENSMUSG00000042472.12 | zinc finger protein 410 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp410 | mm39_v1_chr12_+_84363876_84363920 | -0.77 | 1.3e-01 | Click! |
Activity profile of Zfp410 motif
Sorted Z-values of Zfp410 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_148021217 | 0.14 |
ENSMUST00000019199.14
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr10_+_55982969 | 0.12 |
ENSMUST00000063138.8
|
Msl3l2
|
MSL3 like 2 |
| chr4_-_148021159 | 0.11 |
ENSMUST00000105712.2
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr9_-_49479791 | 0.11 |
ENSMUST00000194252.6
|
Ncam1
|
neural cell adhesion molecule 1 |
| chr5_+_31771281 | 0.10 |
ENSMUST00000031024.14
ENSMUST00000202033.2 |
Mrpl33
Gm43809
|
mitochondrial ribosomal protein L33 predicted gene 43809 |
| chr15_-_79718423 | 0.08 |
ENSMUST00000109623.8
ENSMUST00000109625.8 ENSMUST00000023060.13 ENSMUST00000089299.6 |
Cbx6
Npcd
|
chromobox 6 neuronal pentraxin chromo domain |
| chr5_-_100648487 | 0.07 |
ENSMUST00000239512.1
|
LIN54
|
lin-54 homolog (C. elegans) |
| chr16_-_21606546 | 0.07 |
ENSMUST00000023559.7
|
Ehhadh
|
enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase |
| chr19_+_4905158 | 0.06 |
ENSMUST00000119694.3
ENSMUST00000237504.2 ENSMUST00000237011.2 |
Ctsf
|
cathepsin F |
| chr6_+_125072944 | 0.06 |
ENSMUST00000112392.8
ENSMUST00000056889.15 |
Chd4
|
chromodomain helicase DNA binding protein 4 |
| chr11_+_22462088 | 0.04 |
ENSMUST00000059319.8
|
Tmem17
|
transmembrane protein 17 |
| chr18_-_66135683 | 0.04 |
ENSMUST00000120461.9
ENSMUST00000048260.15 ENSMUST00000236866.2 |
Lman1
|
lectin, mannose-binding, 1 |
| chrX_+_133618693 | 0.03 |
ENSMUST00000113201.8
ENSMUST00000051256.10 ENSMUST00000113199.8 ENSMUST00000035748.14 ENSMUST00000113198.8 ENSMUST00000113197.2 |
Armcx1
|
armadillo repeat containing, X-linked 1 |
| chr6_+_125073108 | 0.03 |
ENSMUST00000112390.8
|
Chd4
|
chromodomain helicase DNA binding protein 4 |
| chr14_-_52434863 | 0.03 |
ENSMUST00000046709.9
|
Supt16
|
SPT16, facilitates chromatin remodeling subunit |
| chr6_+_40419797 | 0.02 |
ENSMUST00000038907.9
ENSMUST00000141490.2 |
Wee2
|
WEE1 homolog 2 (S. pombe) |
| chrX_-_72442342 | 0.01 |
ENSMUST00000180787.3
|
Gm18336
|
predicted gene, 18336 |
| chr5_-_104125226 | 0.01 |
ENSMUST00000048118.15
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr7_+_43865369 | 0.01 |
ENSMUST00000074359.4
|
Klk1b5
|
kallikrein 1-related peptidase b5 |
| chr15_-_79718462 | 0.01 |
ENSMUST00000148358.2
|
Cbx6
|
chromobox 6 |
| chr18_-_44329234 | 0.01 |
ENSMUST00000239421.2
ENSMUST00000097587.5 |
Spink11
|
serine peptidase inhibitor, Kazal type 11 |
| chr13_+_23344776 | 0.01 |
ENSMUST00000228113.2
|
Vmn1r219
|
vomeronasal 1 receptor 219 |
| chr11_-_118931731 | 0.01 |
ENSMUST00000026663.8
|
Cbx8
|
chromobox 8 |
| chr7_+_12712139 | 0.01 |
ENSMUST00000108536.3
|
Zfp446
|
zinc finger protein 446 |
| chr5_-_104125270 | 0.00 |
ENSMUST00000112803.3
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr5_-_104125192 | 0.00 |
ENSMUST00000120320.8
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr7_+_108019628 | 0.00 |
ENSMUST00000213521.3
|
Olfr497
|
olfactory receptor 497 |
| chr15_-_89309998 | 0.00 |
ENSMUST00000168376.2
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr14_-_56299764 | 0.00 |
ENSMUST00000043249.10
|
Mcpt4
|
mast cell protease 4 |
| chr14_-_61495832 | 0.00 |
ENSMUST00000121148.8
|
Sgcg
|
sarcoglycan, gamma (dystrophin-associated glycoprotein) |
| chr5_-_151510389 | 0.00 |
ENSMUST00000165928.4
|
Vmn2r18
|
vomeronasal 2, receptor 18 |
| chr16_+_16888084 | 0.00 |
ENSMUST00000231514.2
|
Ypel1
|
yippee like 1 |
| chr6_-_142418801 | 0.00 |
ENSMUST00000032371.8
|
Gys2
|
glycogen synthase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp410
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |