Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
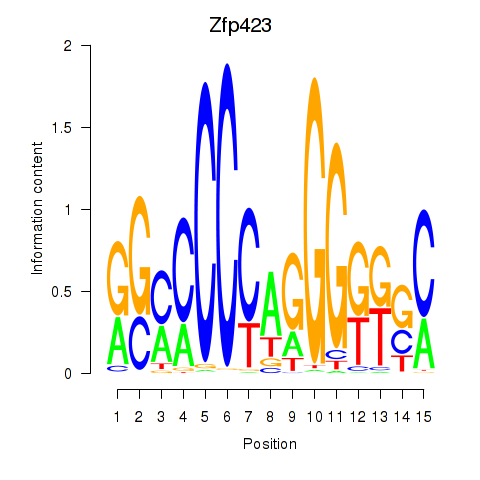
Results for Zfp423
Z-value: 0.11
Transcription factors associated with Zfp423
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp423
|
ENSMUSG00000045333.16 | zinc finger protein 423 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp423 | mm39_v1_chr8_-_88686188_88686223 | -0.55 | 3.3e-01 | Click! |
Activity profile of Zfp423 motif
Sorted Z-values of Zfp423 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_10079091 | 0.12 |
ENSMUST00000025567.9
|
Fads2
|
fatty acid desaturase 2 |
| chr6_-_52217821 | 0.10 |
ENSMUST00000121043.2
|
Hoxa10
|
homeobox A10 |
| chr2_+_154633265 | 0.07 |
ENSMUST00000140713.3
ENSMUST00000137333.8 |
Raly
a
|
hnRNP-associated with lethal yellow nonagouti |
| chr6_-_124391994 | 0.07 |
ENSMUST00000035861.6
ENSMUST00000112532.8 ENSMUST00000080557.12 |
Pex5
|
peroxisomal biogenesis factor 5 |
| chr7_-_120269462 | 0.06 |
ENSMUST00000127845.2
ENSMUST00000208635.2 ENSMUST00000033178.4 |
Pdzd9
|
PDZ domain containing 9 |
| chr2_+_163389068 | 0.05 |
ENSMUST00000109411.8
ENSMUST00000018094.13 |
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chr7_-_126625617 | 0.05 |
ENSMUST00000032916.6
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr2_-_29142965 | 0.05 |
ENSMUST00000155949.2
ENSMUST00000154682.8 ENSMUST00000028141.6 ENSMUST00000071201.5 |
6530402F18Rik
Ntng2
|
RIKEN cDNA 6530402F18 gene netrin G2 |
| chr6_+_7844759 | 0.05 |
ENSMUST00000040159.6
|
C1galt1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr12_-_14202041 | 0.04 |
ENSMUST00000020926.8
|
Lratd1
|
LRAT domain containing 1 |
| chr4_+_129407374 | 0.04 |
ENSMUST00000062356.7
|
Marcksl1
|
MARCKS-like 1 |
| chr19_+_6451667 | 0.04 |
ENSMUST00000113471.3
ENSMUST00000113469.3 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr19_+_10019023 | 0.04 |
ENSMUST00000237672.2
|
Fads3
|
fatty acid desaturase 3 |
| chr4_-_6990774 | 0.04 |
ENSMUST00000039987.4
|
Tox
|
thymocyte selection-associated high mobility group box |
| chr4_-_155845500 | 0.03 |
ENSMUST00000030903.12
|
Atad3a
|
ATPase family, AAA domain containing 3A |
| chr11_-_76737794 | 0.03 |
ENSMUST00000021201.6
|
Cpd
|
carboxypeptidase D |
| chr9_+_120321557 | 0.03 |
ENSMUST00000007139.6
|
Eif1b
|
eukaryotic translation initiation factor 1B |
| chrX_-_74174450 | 0.03 |
ENSMUST00000114092.8
ENSMUST00000132501.8 ENSMUST00000153318.8 ENSMUST00000155742.2 |
Mpp1
|
membrane protein, palmitoylated |
| chr2_-_30249202 | 0.02 |
ENSMUST00000100215.11
ENSMUST00000113620.10 ENSMUST00000163668.3 ENSMUST00000028214.15 ENSMUST00000113621.10 |
Sh3glb2
|
SH3-domain GRB2-like endophilin B2 |
| chr1_+_156386414 | 0.02 |
ENSMUST00000166172.9
ENSMUST00000027888.13 |
Abl2
|
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
| chr6_+_86605146 | 0.02 |
ENSMUST00000043400.9
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr19_+_7245591 | 0.02 |
ENSMUST00000066646.12
|
Rcor2
|
REST corepressor 2 |
| chr2_+_93017887 | 0.02 |
ENSMUST00000150462.8
ENSMUST00000111266.8 |
Trp53i11
|
transformation related protein 53 inducible protein 11 |
| chr3_+_94882038 | 0.02 |
ENSMUST00000072287.12
|
Pi4kb
|
phosphatidylinositol 4-kinase beta |
| chr12_+_102915102 | 0.02 |
ENSMUST00000101099.12
|
Unc79
|
unc-79 homolog |
| chr4_-_148711211 | 0.02 |
ENSMUST00000186947.7
ENSMUST00000105699.8 |
Tardbp
|
TAR DNA binding protein |
| chr11_+_54793743 | 0.02 |
ENSMUST00000149324.3
|
Gpx3
|
glutathione peroxidase 3 |
| chr4_-_116982804 | 0.02 |
ENSMUST00000183310.2
|
Btbd19
|
BTB (POZ) domain containing 19 |
| chr19_+_41818409 | 0.02 |
ENSMUST00000087155.5
|
Frat1
|
frequently rearranged in advanced T cell lymphomas |
| chr19_-_5781503 | 0.02 |
ENSMUST00000162976.2
|
Znrd2
|
zinc ribbon domain containing 2 |
| chr2_+_93017916 | 0.02 |
ENSMUST00000090554.11
|
Trp53i11
|
transformation related protein 53 inducible protein 11 |
| chr10_+_80972368 | 0.02 |
ENSMUST00000119606.8
ENSMUST00000146895.2 ENSMUST00000121840.8 |
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr13_+_41403317 | 0.02 |
ENSMUST00000165561.4
|
Smim13
|
small integral membrane protein 13 |
| chr4_+_126915104 | 0.02 |
ENSMUST00000030623.8
|
Sfpq
|
splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) |
| chr5_-_24628483 | 0.02 |
ENSMUST00000198990.2
|
Cdk5
|
cyclin-dependent kinase 5 |
| chrX_+_100298134 | 0.02 |
ENSMUST00000062000.6
|
Foxo4
|
forkhead box O4 |
| chr4_-_106321363 | 0.01 |
ENSMUST00000049507.6
|
Pcsk9
|
proprotein convertase subtilisin/kexin type 9 |
| chr19_+_10018914 | 0.01 |
ENSMUST00000115995.4
|
Fads3
|
fatty acid desaturase 3 |
| chr13_+_3588063 | 0.01 |
ENSMUST00000223396.2
ENSMUST00000059515.8 ENSMUST00000222365.2 |
Gdi2
|
guanosine diphosphate (GDP) dissociation inhibitor 2 |
| chr7_-_141023199 | 0.01 |
ENSMUST00000106005.9
|
Pidd1
|
p53 induced death domain protein 1 |
| chr7_+_18737944 | 0.01 |
ENSMUST00000053713.5
|
Irf2bp1
|
interferon regulatory factor 2 binding protein 1 |
| chrX_-_74174524 | 0.01 |
ENSMUST00000114091.8
|
Mpp1
|
membrane protein, palmitoylated |
| chr9_-_37464200 | 0.01 |
ENSMUST00000065668.12
|
Nrgn
|
neurogranin |
| chr4_-_148711453 | 0.01 |
ENSMUST00000165113.8
ENSMUST00000172073.8 ENSMUST00000105702.9 ENSMUST00000084125.10 |
Tardbp
|
TAR DNA binding protein |
| chr15_-_98676007 | 0.01 |
ENSMUST00000226655.2
ENSMUST00000228546.2 ENSMUST00000023732.12 ENSMUST00000226610.2 |
Wnt10b
|
wingless-type MMTV integration site family, member 10B |
| chr10_-_14581203 | 0.01 |
ENSMUST00000149485.2
ENSMUST00000154132.8 |
Vta1
|
vesicle (multivesicular body) trafficking 1 |
| chr8_-_41469786 | 0.01 |
ENSMUST00000117735.8
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr7_-_128062916 | 0.01 |
ENSMUST00000205835.2
|
Tial1
|
Tia1 cytotoxic granule-associated RNA binding protein-like 1 |
| chr6_+_113448388 | 0.01 |
ENSMUST00000058300.14
|
Il17rc
|
interleukin 17 receptor C |
| chr5_+_67418137 | 0.01 |
ENSMUST00000161369.3
|
Tmem33
|
transmembrane protein 33 |
| chr7_-_5017642 | 0.01 |
ENSMUST00000207412.2
ENSMUST00000077385.15 ENSMUST00000165320.3 |
Fiz1
|
Flt3 interacting zinc finger protein 1 |
| chr11_+_81992662 | 0.00 |
ENSMUST00000000194.4
|
Ccl12
|
chemokine (C-C motif) ligand 12 |
| chr7_-_19192672 | 0.00 |
ENSMUST00000239292.2
ENSMUST00000085715.7 |
Mark4
|
MAP/microtubule affinity regulating kinase 4 |
| chr15_-_78602313 | 0.00 |
ENSMUST00000229441.2
|
Elfn2
|
leucine rich repeat and fibronectin type III, extracellular 2 |
| chr15_+_78128990 | 0.00 |
ENSMUST00000096357.12
|
Ncf4
|
neutrophil cytosolic factor 4 |
| chr10_+_80973787 | 0.00 |
ENSMUST00000117956.2
|
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr11_-_99228756 | 0.00 |
ENSMUST00000100482.3
|
Krt26
|
keratin 26 |
| chr3_+_114697710 | 0.00 |
ENSMUST00000081752.13
|
Olfm3
|
olfactomedin 3 |
| chr17_-_25652750 | 0.00 |
ENSMUST00000159610.8
ENSMUST00000159048.8 ENSMUST00000078496.12 |
Cacna1h
|
calcium channel, voltage-dependent, T type, alpha 1H subunit |
| chrX_-_74174608 | 0.00 |
ENSMUST00000033775.9
|
Mpp1
|
membrane protein, palmitoylated |
| chr2_+_106523532 | 0.00 |
ENSMUST00000111063.8
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr11_-_43727071 | 0.00 |
ENSMUST00000167574.2
|
Adra1b
|
adrenergic receptor, alpha 1b |
| chr17_-_35293447 | 0.00 |
ENSMUST00000007259.4
|
Ly6g6d
|
lymphocyte antigen 6 complex, locus G6D |
| chr6_+_110622533 | 0.00 |
ENSMUST00000071076.13
ENSMUST00000172951.2 |
Grm7
|
glutamate receptor, metabotropic 7 |
| chr14_+_70791127 | 0.00 |
ENSMUST00000161069.8
|
Hr
|
lysine demethylase and nuclear receptor corepressor |
| chr13_-_114595122 | 0.00 |
ENSMUST00000231252.2
|
Fst
|
follistatin |
| chr9_-_99758200 | 0.00 |
ENSMUST00000054819.10
|
Sox14
|
SRY (sex determining region Y)-box 14 |
| chr16_+_17307503 | 0.00 |
ENSMUST00000023448.15
|
Aifm3
|
apoptosis-inducing factor, mitochondrion-associated 3 |
| chr11_-_33153587 | 0.00 |
ENSMUST00000037746.8
|
Tlx3
|
T cell leukemia, homeobox 3 |
| chr17_-_23939700 | 0.00 |
ENSMUST00000201734.4
|
1520401A03Rik
|
RIKEN cDNA 1520401A03 gene |
| chr17_-_32385826 | 0.00 |
ENSMUST00000087723.5
|
Notch3
|
notch 3 |
| chr15_+_78129040 | 0.00 |
ENSMUST00000133618.3
|
Ncf4
|
neutrophil cytosolic factor 4 |
| chr6_+_6863769 | 0.00 |
ENSMUST00000031768.8
|
Dlx6
|
distal-less homeobox 6 |
| chr16_-_45664664 | 0.00 |
ENSMUST00000036355.13
|
Phldb2
|
pleckstrin homology like domain, family B, member 2 |
| chr4_+_150999019 | 0.00 |
ENSMUST00000135169.8
|
Tnfrsf9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr1_+_167426019 | 0.00 |
ENSMUST00000111386.8
ENSMUST00000111384.8 |
Rxrg
|
retinoid X receptor gamma |
| chr15_-_98675142 | 0.00 |
ENSMUST00000166022.2
|
Wnt10b
|
wingless-type MMTV integration site family, member 10B |
| chr11_+_102727122 | 0.00 |
ENSMUST00000021302.15
ENSMUST00000107072.2 |
Higd1b
|
HIG1 domain family, member 1B |
| chr2_+_180141686 | 0.00 |
ENSMUST00000029084.9
|
Ntsr1
|
neurotensin receptor 1 |
| chr4_+_154321982 | 0.00 |
ENSMUST00000152159.8
|
Megf6
|
multiple EGF-like-domains 6 |
| chr18_-_66072858 | 0.00 |
ENSMUST00000237506.2
|
Rax
|
retina and anterior neural fold homeobox |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp423
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.0 | 0.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.1 | GO:1902569 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.0 | 0.1 | GO:0031782 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |