Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
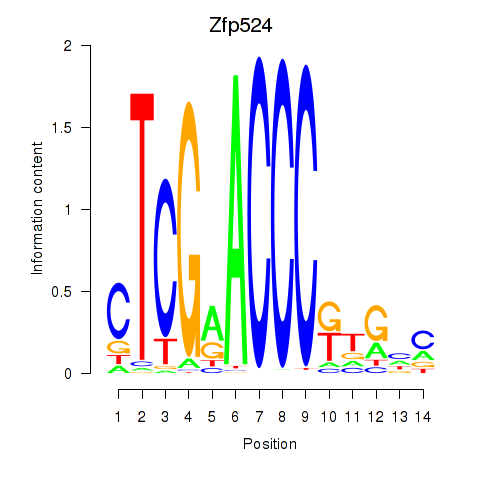
Results for Zfp524
Z-value: 0.95
Transcription factors associated with Zfp524
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp524
|
ENSMUSG00000051184.8 | zinc finger protein 524 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp524 | mm39_v1_chr7_+_5018453_5018509 | -0.35 | 5.6e-01 | Click! |
Activity profile of Zfp524 motif
Sorted Z-values of Zfp524 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_98247237 | 0.69 |
ENSMUST00000065793.12
|
Phgdh
|
3-phosphoglycerate dehydrogenase |
| chr4_-_120427449 | 0.61 |
ENSMUST00000030381.8
|
Ctps
|
cytidine 5'-triphosphate synthase |
| chr8_+_71858647 | 0.57 |
ENSMUST00000119976.8
ENSMUST00000120725.2 |
Ankle1
|
ankyrin repeat and LEM domain containing 1 |
| chr12_-_75678092 | 0.51 |
ENSMUST00000238938.2
|
Rplp2-ps1
|
ribosomal protein, large P2, pseudogene 1 |
| chr15_-_79967543 | 0.44 |
ENSMUST00000081650.15
|
Rpl3
|
ribosomal protein L3 |
| chr16_+_35759346 | 0.41 |
ENSMUST00000023622.13
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr12_+_112611322 | 0.38 |
ENSMUST00000109755.5
|
Siva1
|
SIVA1, apoptosis-inducing factor |
| chr13_-_96807346 | 0.38 |
ENSMUST00000022176.15
|
Hmgcr
|
3-hydroxy-3-methylglutaryl-Coenzyme A reductase |
| chr16_+_16714333 | 0.38 |
ENSMUST00000027373.12
ENSMUST00000232247.2 |
Ppm1f
|
protein phosphatase 1F (PP2C domain containing) |
| chr5_-_23988551 | 0.37 |
ENSMUST00000148618.8
|
Pus7
|
pseudouridylate synthase 7 |
| chr7_-_30428746 | 0.35 |
ENSMUST00000209065.2
ENSMUST00000208169.2 |
Tmem147
|
transmembrane protein 147 |
| chr19_+_45549009 | 0.35 |
ENSMUST00000047057.9
|
Gm17018
|
predicted gene 17018 |
| chr13_+_8935537 | 0.34 |
ENSMUST00000169314.9
|
Idi1
|
isopentenyl-diphosphate delta isomerase |
| chr13_+_34221572 | 0.34 |
ENSMUST00000040656.8
|
Bphl
|
biphenyl hydrolase-like (serine hydrolase, breast epithelial mucin-associated antigen) |
| chr15_-_36496880 | 0.34 |
ENSMUST00000228601.2
ENSMUST00000057486.9 |
Ankrd46
|
ankyrin repeat domain 46 |
| chr11_-_6217718 | 0.34 |
ENSMUST00000004507.11
ENSMUST00000151446.2 |
Ddx56
|
DEAD box helicase 56 |
| chr4_-_136626073 | 0.33 |
ENSMUST00000046285.6
|
C1qa
|
complement component 1, q subcomponent, alpha polypeptide |
| chr8_-_111629074 | 0.33 |
ENSMUST00000041382.7
|
Fcsk
|
fucose kinase |
| chr16_-_4536992 | 0.33 |
ENSMUST00000115851.10
|
Nmral1
|
NmrA-like family domain containing 1 |
| chr4_+_131570770 | 0.33 |
ENSMUST00000030742.11
ENSMUST00000137321.3 |
Mecr
|
mitochondrial trans-2-enoyl-CoA reductase |
| chr5_+_115039359 | 0.33 |
ENSMUST00000124716.3
|
Oasl2
|
2'-5' oligoadenylate synthetase-like 2 |
| chr3_-_89009153 | 0.33 |
ENSMUST00000199668.3
ENSMUST00000196709.5 |
Fdps
|
farnesyl diphosphate synthetase |
| chr17_-_26080429 | 0.33 |
ENSMUST00000079461.15
ENSMUST00000176923.9 |
Wdr90
|
WD repeat domain 90 |
| chr5_+_28276353 | 0.31 |
ENSMUST00000059155.11
|
Insig1
|
insulin induced gene 1 |
| chr13_-_6686686 | 0.31 |
ENSMUST00000136585.2
|
Pfkp
|
phosphofructokinase, platelet |
| chr10_+_81093110 | 0.31 |
ENSMUST00000117488.8
ENSMUST00000105328.10 ENSMUST00000121205.8 |
Matk
|
megakaryocyte-associated tyrosine kinase |
| chr12_+_30634425 | 0.30 |
ENSMUST00000057151.10
|
Tmem18
|
transmembrane protein 18 |
| chr5_-_23988696 | 0.30 |
ENSMUST00000119946.8
|
Pus7
|
pseudouridylate synthase 7 |
| chr7_+_24069680 | 0.29 |
ENSMUST00000205428.2
ENSMUST00000171904.3 ENSMUST00000205626.2 |
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr4_-_129229159 | 0.29 |
ENSMUST00000102598.4
|
Rbbp4
|
retinoblastoma binding protein 4, chromatin remodeling factor |
| chr7_+_4467730 | 0.28 |
ENSMUST00000086372.8
ENSMUST00000169820.8 ENSMUST00000163893.8 ENSMUST00000170635.2 |
Eps8l1
|
EPS8-like 1 |
| chr7_+_127345909 | 0.28 |
ENSMUST00000033081.14
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr7_-_30428930 | 0.28 |
ENSMUST00000207296.2
ENSMUST00000006478.10 |
Tmem147
|
transmembrane protein 147 |
| chr3_-_94794256 | 0.28 |
ENSMUST00000005923.7
|
Psmb4
|
proteasome (prosome, macropain) subunit, beta type 4 |
| chr3_+_97066065 | 0.28 |
ENSMUST00000090759.5
|
Acp6
|
acid phosphatase 6, lysophosphatidic |
| chr10_+_11157326 | 0.27 |
ENSMUST00000070300.5
|
Fbxo30
|
F-box protein 30 |
| chr7_+_28524627 | 0.26 |
ENSMUST00000066264.13
|
Ech1
|
enoyl coenzyme A hydratase 1, peroxisomal |
| chr6_+_137731599 | 0.26 |
ENSMUST00000204356.2
|
Dera
|
deoxyribose-phosphate aldolase (putative) |
| chr16_+_32427738 | 0.25 |
ENSMUST00000023486.15
|
Tfrc
|
transferrin receptor |
| chr3_-_89009214 | 0.25 |
ENSMUST00000081848.13
|
Fdps
|
farnesyl diphosphate synthetase |
| chr5_-_74229021 | 0.25 |
ENSMUST00000119154.8
ENSMUST00000068058.14 |
Usp46
|
ubiquitin specific peptidase 46 |
| chr13_+_54225828 | 0.25 |
ENSMUST00000021930.10
|
Sfxn1
|
sideroflexin 1 |
| chr7_-_133378410 | 0.24 |
ENSMUST00000130182.2
ENSMUST00000106139.8 |
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr15_+_76215488 | 0.24 |
ENSMUST00000172281.8
|
Gpaa1
|
GPI anchor attachment protein 1 |
| chr19_+_8944369 | 0.24 |
ENSMUST00000052248.8
|
Eef1g
|
eukaryotic translation elongation factor 1 gamma |
| chr19_-_42741148 | 0.24 |
ENSMUST00000236659.2
ENSMUST00000076505.4 |
Pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr18_+_12732951 | 0.23 |
ENSMUST00000234255.2
ENSMUST00000169401.8 |
Ttc39c
|
tetratricopeptide repeat domain 39C |
| chr5_+_52898910 | 0.23 |
ENSMUST00000031081.11
ENSMUST00000031082.8 |
Pi4k2b
|
phosphatidylinositol 4-kinase type 2 beta |
| chr8_-_11600689 | 0.23 |
ENSMUST00000049461.7
|
Cars2
|
cysteinyl-tRNA synthetase 2 (mitochondrial)(putative) |
| chr3_+_69629318 | 0.23 |
ENSMUST00000029358.15
|
Nmd3
|
NMD3 ribosome export adaptor |
| chr7_-_46365108 | 0.23 |
ENSMUST00000006956.9
ENSMUST00000210913.2 |
Saa3
|
serum amyloid A 3 |
| chr15_+_44291470 | 0.23 |
ENSMUST00000226827.2
ENSMUST00000060652.5 |
Eny2
|
ENY2 transcription and export complex 2 subunit |
| chr11_-_74614654 | 0.22 |
ENSMUST00000102520.9
|
Pafah1b1
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 1 |
| chr4_-_149221998 | 0.22 |
ENSMUST00000176124.8
ENSMUST00000177408.2 ENSMUST00000105695.2 |
Cenps
|
centromere protein S |
| chr13_-_96807326 | 0.22 |
ENSMUST00000169196.8
|
Hmgcr
|
3-hydroxy-3-methylglutaryl-Coenzyme A reductase |
| chr7_-_132178101 | 0.22 |
ENSMUST00000084500.8
|
Oat
|
ornithine aminotransferase |
| chr2_-_3513783 | 0.22 |
ENSMUST00000124331.8
ENSMUST00000140494.2 ENSMUST00000027961.12 |
Gm45902
Hspa14
|
predicted gene 45902 heat shock protein 14 |
| chr19_+_10554799 | 0.22 |
ENSMUST00000237564.2
ENSMUST00000236743.2 ENSMUST00000235271.2 ENSMUST00000168445.2 ENSMUST00000237641.2 ENSMUST00000236352.2 |
Cyb561a3
|
cytochrome b561 family, member A3 |
| chr4_-_149222057 | 0.22 |
ENSMUST00000030813.10
|
Cenps
|
centromere protein S |
| chr15_+_76763501 | 0.22 |
ENSMUST00000230106.2
ENSMUST00000229831.2 ENSMUST00000229990.2 ENSMUST00000230214.2 |
Zfp7
Gm49527
|
zinc finger protein 7 predicted gene, 49527 |
| chr11_+_109540201 | 0.22 |
ENSMUST00000106677.8
|
Prkar1a
|
protein kinase, cAMP dependent regulatory, type I, alpha |
| chr17_+_34062059 | 0.22 |
ENSMUST00000002379.15
|
Cd320
|
CD320 antigen |
| chr12_-_71183371 | 0.21 |
ENSMUST00000221367.2
ENSMUST00000220482.2 ENSMUST00000221892.2 ENSMUST00000221178.2 ENSMUST00000221559.2 ENSMUST00000166120.9 ENSMUST00000021486.10 ENSMUST00000221797.2 ENSMUST00000221815.2 |
Timm9
|
translocase of inner mitochondrial membrane 9 |
| chrX_-_168103266 | 0.21 |
ENSMUST00000033717.9
ENSMUST00000112115.2 |
Hccs
|
holocytochrome c synthetase |
| chr7_+_15853792 | 0.21 |
ENSMUST00000006178.5
|
Kptn
|
kaptin |
| chr16_+_35758836 | 0.21 |
ENSMUST00000114878.8
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr2_-_132095146 | 0.21 |
ENSMUST00000028817.7
|
Pcna
|
proliferating cell nuclear antigen |
| chr2_+_181023028 | 0.20 |
ENSMUST00000048077.12
|
Lime1
|
Lck interacting transmembrane adaptor 1 |
| chr8_-_71938598 | 0.20 |
ENSMUST00000093450.6
ENSMUST00000213382.2 |
Ano8
|
anoctamin 8 |
| chr14_+_8508484 | 0.20 |
ENSMUST00000065865.10
ENSMUST00000225891.2 |
Thoc7
|
THO complex 7 |
| chr14_+_69585036 | 0.20 |
ENSMUST00000064831.6
|
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr7_+_78545660 | 0.20 |
ENSMUST00000107425.8
ENSMUST00000107421.8 |
Aen
|
apoptosis enhancing nuclease |
| chr12_-_107969673 | 0.20 |
ENSMUST00000109887.8
ENSMUST00000109891.3 |
Bcl11b
|
B cell leukemia/lymphoma 11B |
| chr3_+_87814147 | 0.20 |
ENSMUST00000159492.8
|
Hdgf
|
heparin binding growth factor |
| chr7_+_78545756 | 0.20 |
ENSMUST00000107423.2
|
Aen
|
apoptosis enhancing nuclease |
| chr17_+_29312737 | 0.20 |
ENSMUST00000023829.8
ENSMUST00000233296.2 |
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr1_-_75187417 | 0.19 |
ENSMUST00000113623.8
|
Glb1l
|
galactosidase, beta 1-like |
| chr16_-_4536943 | 0.19 |
ENSMUST00000120056.8
ENSMUST00000074970.8 |
Nmral1
|
NmrA-like family domain containing 1 |
| chr9_-_106315518 | 0.19 |
ENSMUST00000024031.13
ENSMUST00000190972.3 |
Acy1
|
aminoacylase 1 |
| chr17_+_6307123 | 0.19 |
ENSMUST00000232383.2
|
Tmem181a
|
transmembrane protein 181A |
| chr2_+_103800459 | 0.19 |
ENSMUST00000111143.8
ENSMUST00000138815.2 |
Lmo2
|
LIM domain only 2 |
| chr6_-_56681657 | 0.19 |
ENSMUST00000176595.3
ENSMUST00000170382.5 ENSMUST00000203958.2 |
Lsm5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr11_+_5048915 | 0.19 |
ENSMUST00000101610.10
|
Rhbdd3
|
rhomboid domain containing 3 |
| chr8_+_105067159 | 0.19 |
ENSMUST00000212948.2
ENSMUST00000034343.5 |
Cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr11_+_69792642 | 0.18 |
ENSMUST00000177138.8
ENSMUST00000108617.10 ENSMUST00000177476.8 ENSMUST00000061837.11 |
Neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr8_-_73059104 | 0.18 |
ENSMUST00000075602.8
|
Gm10282
|
predicted pseudogene 10282 |
| chr12_-_110662677 | 0.18 |
ENSMUST00000124156.8
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr2_+_90507552 | 0.18 |
ENSMUST00000057481.7
|
Nup160
|
nucleoporin 160 |
| chr5_+_33815466 | 0.18 |
ENSMUST00000074849.13
ENSMUST00000079534.11 ENSMUST00000201633.2 |
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr8_-_124747747 | 0.18 |
ENSMUST00000093039.6
|
Taf5l
|
TATA-box binding protein associated factor 5 like |
| chr13_-_63579497 | 0.18 |
ENSMUST00000160931.2
ENSMUST00000099444.10 ENSMUST00000220684.2 ENSMUST00000161977.8 ENSMUST00000163091.8 |
Fancc
|
Fanconi anemia, complementation group C |
| chr11_+_23615612 | 0.18 |
ENSMUST00000109525.8
ENSMUST00000020520.11 |
Pus10
|
pseudouridylate synthase 10 |
| chr6_+_137731526 | 0.18 |
ENSMUST00000203216.3
ENSMUST00000087675.9 ENSMUST00000203693.3 |
Dera
|
deoxyribose-phosphate aldolase (putative) |
| chr9_-_114610879 | 0.18 |
ENSMUST00000084867.9
ENSMUST00000216760.2 ENSMUST00000035009.16 |
Cmtm7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr19_-_55304392 | 0.18 |
ENSMUST00000224291.2
ENSMUST00000225495.2 ENSMUST00000076891.7 ENSMUST00000224897.2 |
Zdhhc6
|
zinc finger, DHHC domain containing 6 |
| chr7_-_120581535 | 0.18 |
ENSMUST00000033169.9
|
Cdr2
|
cerebellar degeneration-related 2 |
| chr19_+_10554510 | 0.18 |
ENSMUST00000237814.2
|
Cyb561a3
|
cytochrome b561 family, member A3 |
| chr6_+_127423779 | 0.17 |
ENSMUST00000112191.8
|
Parp11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr8_+_80366247 | 0.17 |
ENSMUST00000173078.8
ENSMUST00000173286.8 |
Otud4
|
OTU domain containing 4 |
| chr3_+_32763313 | 0.17 |
ENSMUST00000126144.3
|
Actl6a
|
actin-like 6A |
| chr19_-_45548942 | 0.16 |
ENSMUST00000026239.7
|
Poll
|
polymerase (DNA directed), lambda |
| chr7_-_126483851 | 0.16 |
ENSMUST00000071268.11
ENSMUST00000117394.2 |
Taok2
|
TAO kinase 2 |
| chr15_-_12824691 | 0.16 |
ENSMUST00000228177.2
ENSMUST00000227299.2 ENSMUST00000057256.5 |
6030458C11Rik
|
RIKEN cDNA 6030458C11 gene |
| chr2_+_103799873 | 0.16 |
ENSMUST00000123437.8
|
Lmo2
|
LIM domain only 2 |
| chr11_+_62770275 | 0.16 |
ENSMUST00000014321.5
|
Tvp23b
|
trans-golgi network vesicle protein 23B |
| chr15_+_103411689 | 0.16 |
ENSMUST00000226493.2
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent |
| chr13_-_99027544 | 0.16 |
ENSMUST00000109399.9
|
Tnpo1
|
transportin 1 |
| chr5_-_137531463 | 0.16 |
ENSMUST00000170293.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr5_+_38417710 | 0.16 |
ENSMUST00000119047.8
|
Tmem128
|
transmembrane protein 128 |
| chrX_+_149330371 | 0.16 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr12_+_72807985 | 0.16 |
ENSMUST00000021514.10
|
Ppm1a
|
protein phosphatase 1A, magnesium dependent, alpha isoform |
| chr17_-_46343291 | 0.16 |
ENSMUST00000071648.12
ENSMUST00000142351.9 |
Vegfa
|
vascular endothelial growth factor A |
| chr8_+_72044802 | 0.16 |
ENSMUST00000034264.11
|
Pgls
|
6-phosphogluconolactonase |
| chr5_-_100867520 | 0.16 |
ENSMUST00000112908.2
ENSMUST00000045617.15 |
Hpse
|
heparanase |
| chr9_-_75316625 | 0.15 |
ENSMUST00000168937.8
|
Mapk6
|
mitogen-activated protein kinase 6 |
| chr19_+_40819682 | 0.15 |
ENSMUST00000025983.13
ENSMUST00000119316.2 |
Ccnj
|
cyclin J |
| chr2_+_181023120 | 0.15 |
ENSMUST00000126611.8
|
Lime1
|
Lck interacting transmembrane adaptor 1 |
| chr14_+_8507874 | 0.15 |
ENSMUST00000225325.2
|
Thoc7
|
THO complex 7 |
| chr8_-_27618643 | 0.15 |
ENSMUST00000033877.6
|
Brf2
|
BRF2, RNA polymerase III transcription initiation factor 50kDa subunit |
| chr17_+_74796473 | 0.15 |
ENSMUST00000024873.7
|
Yipf4
|
Yip1 domain family, member 4 |
| chr14_+_55815879 | 0.15 |
ENSMUST00000174563.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr3_+_40699763 | 0.15 |
ENSMUST00000203353.3
|
Hspa4l
|
heat shock protein 4 like |
| chr6_-_87827993 | 0.15 |
ENSMUST00000204890.3
ENSMUST00000113617.3 ENSMUST00000113619.8 ENSMUST00000204653.3 ENSMUST00000032138.15 |
Cnbp
|
cellular nucleic acid binding protein |
| chr17_+_79223946 | 0.14 |
ENSMUST00000063817.7
|
Cebpzos
|
CCAAT/enhancer binding protein (C/EBP), zeta, opposite strand |
| chr8_-_124748124 | 0.14 |
ENSMUST00000165628.9
|
Taf5l
|
TATA-box binding protein associated factor 5 like |
| chr15_+_76215431 | 0.14 |
ENSMUST00000023221.13
|
Gpaa1
|
GPI anchor attachment protein 1 |
| chr11_+_78234300 | 0.14 |
ENSMUST00000002127.14
ENSMUST00000108295.8 |
Unc119
|
unc-119 lipid binding chaperone |
| chr8_+_96058907 | 0.14 |
ENSMUST00000034245.16
|
Usb1
|
U6 snRNA biogenesis 1 |
| chr2_-_144112700 | 0.14 |
ENSMUST00000110030.10
|
Snx5
|
sorting nexin 5 |
| chr18_-_36877571 | 0.14 |
ENSMUST00000014438.5
|
Ndufa2
|
NADH:ubiquinone oxidoreductase subunit A2 |
| chr13_-_107073415 | 0.14 |
ENSMUST00000080856.14
|
Ipo11
|
importin 11 |
| chr14_+_56640038 | 0.14 |
ENSMUST00000095793.3
ENSMUST00000223627.2 |
Rnf17
|
ring finger protein 17 |
| chr12_+_110816738 | 0.14 |
ENSMUST00000222460.2
|
Zfp839
|
zinc finger protein 839 |
| chr8_-_13940234 | 0.14 |
ENSMUST00000033839.9
|
Coprs
|
coordinator of PRMT5, differentiation stimulator |
| chr9_-_57552392 | 0.14 |
ENSMUST00000216934.2
|
Csk
|
c-src tyrosine kinase |
| chr11_-_73067828 | 0.13 |
ENSMUST00000108480.2
ENSMUST00000054952.4 |
Emc6
|
ER membrane protein complex subunit 6 |
| chr11_+_105927698 | 0.13 |
ENSMUST00000058438.9
|
Dcaf7
|
DDB1 and CUL4 associated factor 7 |
| chr10_-_80742244 | 0.13 |
ENSMUST00000105332.3
|
Lmnb2
|
lamin B2 |
| chr15_+_102204691 | 0.13 |
ENSMUST00000064924.6
ENSMUST00000230212.2 ENSMUST00000229050.2 ENSMUST00000231104.2 |
Espl1
|
extra spindle pole bodies 1, separase |
| chr8_-_13939964 | 0.13 |
ENSMUST00000209371.2
|
Coprs
|
coordinator of PRMT5, differentiation stimulator |
| chr11_-_20282684 | 0.13 |
ENSMUST00000004634.7
ENSMUST00000109594.8 |
Slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr11_+_5049404 | 0.13 |
ENSMUST00000134267.8
ENSMUST00000036320.12 ENSMUST00000150632.8 |
Rhbdd3
|
rhomboid domain containing 3 |
| chr7_-_126074256 | 0.13 |
ENSMUST00000205440.2
ENSMUST00000032978.8 ENSMUST00000205340.2 |
Sh2b1
|
SH2B adaptor protein 1 |
| chr7_-_30262337 | 0.13 |
ENSMUST00000207031.2
|
Psenen
|
presenilin enhancer gamma secretase subunit |
| chr6_+_38511758 | 0.13 |
ENSMUST00000019833.5
|
Fmc1
|
formation of mitochondrial complex V assembly factor 1 |
| chr4_+_155334673 | 0.13 |
ENSMUST00000143709.2
|
Faap20
|
Fanconi anemia core complex associated protein 20 |
| chr17_+_8502682 | 0.13 |
ENSMUST00000124023.8
|
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr1_+_58432629 | 0.13 |
ENSMUST00000186949.2
|
Bzw1
|
basic leucine zipper and W2 domains 1 |
| chr5_-_114518566 | 0.13 |
ENSMUST00000102581.11
|
Kctd10
|
potassium channel tetramerisation domain containing 10 |
| chr17_+_45997248 | 0.13 |
ENSMUST00000024734.8
|
Mrpl14
|
mitochondrial ribosomal protein L14 |
| chr13_-_58532990 | 0.13 |
ENSMUST00000022032.7
|
2210016F16Rik
|
RIKEN cDNA 2210016F16 gene |
| chr14_-_77274056 | 0.13 |
ENSMUST00000062789.15
|
Lacc1
|
laccase domain containing 1 |
| chr3_+_89366425 | 0.12 |
ENSMUST00000029564.12
|
Pmvk
|
phosphomevalonate kinase |
| chr7_-_24672055 | 0.12 |
ENSMUST00000205871.2
|
Rabac1
|
Rab acceptor 1 (prenylated) |
| chr5_+_65264863 | 0.12 |
ENSMUST00000204097.3
|
Klhl5
|
kelch-like 5 |
| chr4_+_149671012 | 0.12 |
ENSMUST00000039144.7
|
Clstn1
|
calsyntenin 1 |
| chr16_-_87229485 | 0.12 |
ENSMUST00000039449.9
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chr11_+_55360502 | 0.12 |
ENSMUST00000018727.4
|
G3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr2_-_24653059 | 0.12 |
ENSMUST00000100348.10
ENSMUST00000041342.12 ENSMUST00000114447.8 ENSMUST00000102939.9 ENSMUST00000070864.14 |
Cacna1b
|
calcium channel, voltage-dependent, N type, alpha 1B subunit |
| chr5_+_33815910 | 0.12 |
ENSMUST00000114426.10
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chrX_-_139857424 | 0.12 |
ENSMUST00000033805.15
ENSMUST00000112978.2 |
Psmd10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr17_-_25069263 | 0.12 |
ENSMUST00000049642.6
|
Fahd1
|
fumarylacetoacetate hydrolase domain containing 1 |
| chr8_+_105066924 | 0.12 |
ENSMUST00000212081.2
|
Cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr10_+_13376745 | 0.12 |
ENSMUST00000060212.13
ENSMUST00000121465.3 |
Fuca2
|
fucosidase, alpha-L- 2, plasma |
| chr7_+_125043806 | 0.12 |
ENSMUST00000033010.9
ENSMUST00000135129.2 |
Kdm8
|
lysine (K)-specific demethylase 8 |
| chr1_+_75187474 | 0.12 |
ENSMUST00000027401.11
ENSMUST00000144355.8 ENSMUST00000123825.8 ENSMUST00000189698.7 |
Stk16
|
serine/threonine kinase 16 |
| chrX_-_70536449 | 0.12 |
ENSMUST00000037391.12
ENSMUST00000114586.9 ENSMUST00000114587.3 |
Cd99l2
|
CD99 antigen-like 2 |
| chr13_-_34119937 | 0.12 |
ENSMUST00000170991.8
ENSMUST00000171252.8 ENSMUST00000164627.8 ENSMUST00000017188.14 ENSMUST00000167163.8 ENSMUST00000043552.17 |
Serpinb6a
|
serine (or cysteine) peptidase inhibitor, clade B, member 6a |
| chr11_-_68899248 | 0.11 |
ENSMUST00000021282.12
|
Pfas
|
phosphoribosylformylglycinamidine synthase (FGAR amidotransferase) |
| chr3_-_105594865 | 0.11 |
ENSMUST00000090680.11
|
Ddx20
|
DEAD box helicase 20 |
| chr2_+_71617402 | 0.11 |
ENSMUST00000238991.2
|
Itga6
|
integrin alpha 6 |
| chr18_+_67221287 | 0.11 |
ENSMUST00000025402.15
|
Gnal
|
guanine nucleotide binding protein, alpha stimulating, olfactory type |
| chr9_-_62719208 | 0.11 |
ENSMUST00000034775.10
|
Fem1b
|
fem 1 homolog b |
| chr2_+_122065230 | 0.11 |
ENSMUST00000110551.4
|
Sord
|
sorbitol dehydrogenase |
| chr7_-_126073994 | 0.11 |
ENSMUST00000205733.2
ENSMUST00000205889.2 |
Sh2b1
|
SH2B adaptor protein 1 |
| chr2_-_144112444 | 0.11 |
ENSMUST00000028909.5
|
Snx5
|
sorting nexin 5 |
| chr19_+_10554845 | 0.11 |
ENSMUST00000237581.2
|
Cyb561a3
|
cytochrome b561 family, member A3 |
| chr7_-_141014477 | 0.11 |
ENSMUST00000106007.10
ENSMUST00000150026.2 ENSMUST00000202840.4 ENSMUST00000133206.9 |
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr4_+_130253925 | 0.11 |
ENSMUST00000105994.4
|
Snrnp40
|
small nuclear ribonucleoprotein 40 (U5) |
| chrX_-_56371953 | 0.11 |
ENSMUST00000176986.8
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr7_-_19363280 | 0.11 |
ENSMUST00000094762.10
ENSMUST00000049912.15 ENSMUST00000098754.5 |
Relb
|
avian reticuloendotheliosis viral (v-rel) oncogene related B |
| chr10_+_80062468 | 0.11 |
ENSMUST00000130260.2
|
Pwwp3a
|
PWWP domain containing 3A, DNA repair factor |
| chr15_+_6451721 | 0.11 |
ENSMUST00000163082.2
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr7_+_101732323 | 0.11 |
ENSMUST00000124189.3
|
Trpc2
|
transient receptor potential cation channel, subfamily C, member 2 |
| chr7_-_126074222 | 0.11 |
ENSMUST00000205497.2
|
Sh2b1
|
SH2B adaptor protein 1 |
| chr1_+_86514822 | 0.11 |
ENSMUST00000027446.11
|
Cops7b
|
COP9 signalosome subunit 7B |
| chr1_+_162398084 | 0.11 |
ENSMUST00000132158.8
ENSMUST00000135241.8 |
Vamp4
|
vesicle-associated membrane protein 4 |
| chr7_-_19005721 | 0.11 |
ENSMUST00000032561.9
|
Vasp
|
vasodilator-stimulated phosphoprotein |
| chr7_-_45083688 | 0.11 |
ENSMUST00000210439.2
|
Ruvbl2
|
RuvB-like protein 2 |
| chr6_-_86710250 | 0.11 |
ENSMUST00000001185.14
|
Gmcl1
|
germ cell-less, spermatogenesis associated 1 |
| chr9_+_64080644 | 0.10 |
ENSMUST00000034966.9
|
Rpl4
|
ribosomal protein L4 |
| chr8_+_72044873 | 0.10 |
ENSMUST00000138742.8
ENSMUST00000143441.2 |
Pgls
|
6-phosphogluconolactonase |
| chr16_-_18161746 | 0.10 |
ENSMUST00000231372.2
ENSMUST00000130752.2 ENSMUST00000231605.2 ENSMUST00000115628.10 |
Tango2
|
transport and golgi organization 2 |
| chr8_-_121394742 | 0.10 |
ENSMUST00000181334.2
ENSMUST00000181950.2 ENSMUST00000181333.2 |
Emc8
Gm27021
|
ER membrane protein complex subunit 8 predicted gene, 27021 |
| chrX_-_70536198 | 0.10 |
ENSMUST00000080035.11
|
Cd99l2
|
CD99 antigen-like 2 |
| chr9_-_50515089 | 0.10 |
ENSMUST00000000175.6
|
Sdhd
|
succinate dehydrogenase complex, subunit D, integral membrane protein |
| chr19_+_21630540 | 0.10 |
ENSMUST00000235332.2
|
Abhd17b
|
abhydrolase domain containing 17B |
| chr12_-_102724931 | 0.10 |
ENSMUST00000174651.2
|
Gm20604
|
predicted gene 20604 |
| chr7_+_4743114 | 0.10 |
ENSMUST00000098853.9
ENSMUST00000130215.8 ENSMUST00000108582.10 ENSMUST00000108583.9 |
Kmt5c
|
lysine methyltransferase 5C |
| chr2_+_31560725 | 0.10 |
ENSMUST00000038474.14
ENSMUST00000137156.2 |
Exosc2
|
exosome component 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp524
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.1 | 0.7 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.1 | 0.6 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.1 | 0.4 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.1 | 0.4 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 0.3 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.3 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 | 0.2 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 0.3 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 0.2 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.1 | 0.4 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 0.3 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 | 0.3 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.2 | GO:0038189 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.1 | 0.2 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.2 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.3 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 | 0.6 | GO:0046036 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.8 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.3 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) alditol catabolic process(GO:0019405) |
| 0.0 | 0.4 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.6 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.0 | 0.1 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.5 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.8 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.2 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.4 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.3 | GO:0036005 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.0 | 0.1 | GO:0090579 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.3 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.2 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:0009726 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.4 | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901030) |
| 0.0 | 0.1 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.4 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.3 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 0.0 | 0.0 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.1 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.0 | 0.1 | GO:0090034 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.3 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.1 | GO:0090003 | regulation of Golgi to plasma membrane protein transport(GO:0042996) regulation of establishment of protein localization to plasma membrane(GO:0090003) |
| 0.0 | 0.0 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 0.0 | 0.1 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.3 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.3 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.0 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.0 | GO:0046108 | uridine catabolic process(GO:0006218) uridine metabolic process(GO:0046108) |
| 0.0 | 0.0 | GO:0071874 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.7 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.2 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.0 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.2 | GO:0072431 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.0 | 0.1 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.1 | GO:0051307 | meiotic chromosome separation(GO:0051307) |
| 0.0 | 0.0 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.0 | 0.1 | GO:1905161 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.0 | 0.1 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.4 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 0.3 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.4 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 0.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.3 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.0 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.1 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.0 | 0.1 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.6 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.1 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0034680 | integrin alpha1-beta1 complex(GO:0034665) integrin alpha7-beta1 complex(GO:0034677) integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.0 | GO:1990622 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.0 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.1 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.0 | 0.0 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0042282 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 0.2 | 0.6 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.6 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.3 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.1 | 0.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.3 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 0.3 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.1 | 0.2 | GO:0032139 | DNA polymerase processivity factor activity(GO:0030337) dinucleotide insertion or deletion binding(GO:0032139) |
| 0.1 | 0.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 0.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.4 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.2 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.3 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.1 | 0.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.3 | GO:0002135 | CTP binding(GO:0002135) |
| 0.0 | 0.1 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.1 | GO:0016823 | oxaloacetate decarboxylase activity(GO:0008948) hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.3 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.2 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.0 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.0 | 0.3 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.0 | 0.4 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.4 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.1 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0015193 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.3 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.6 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.2 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |