Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
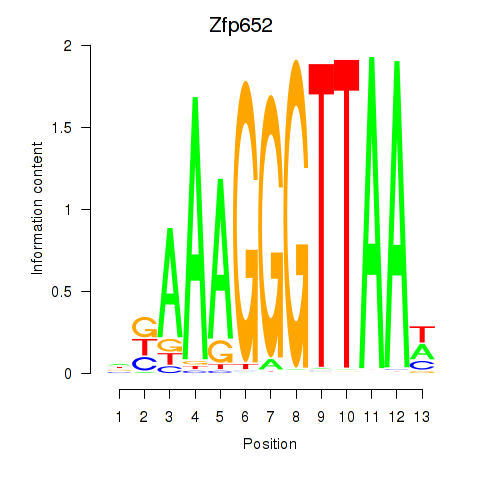
Results for Zfp652
Z-value: 1.18
Transcription factors associated with Zfp652
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp652
|
ENSMUSG00000075595.10 | zinc finger protein 652 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp652 | mm39_v1_chr11_+_95603494_95603529 | 0.89 | 4.4e-02 | Click! |
Activity profile of Zfp652 motif
Sorted Z-values of Zfp652 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_94932158 | 1.22 |
ENSMUST00000038431.8
|
Pdk2
|
pyruvate dehydrogenase kinase, isoenzyme 2 |
| chr11_+_98851238 | 1.09 |
ENSMUST00000107473.3
|
Rara
|
retinoic acid receptor, alpha |
| chr11_+_45946800 | 0.99 |
ENSMUST00000011400.8
|
Adam19
|
a disintegrin and metallopeptidase domain 19 (meltrin beta) |
| chr19_-_30526916 | 0.82 |
ENSMUST00000025803.9
|
Dkk1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr4_-_63321591 | 0.70 |
ENSMUST00000035724.5
|
Akna
|
AT-hook transcription factor |
| chr1_+_143516402 | 0.66 |
ENSMUST00000038252.4
|
B3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr9_+_75682637 | 0.66 |
ENSMUST00000012281.8
|
Bmp5
|
bone morphogenetic protein 5 |
| chrX_+_109857866 | 0.63 |
ENSMUST00000078229.5
|
Pou3f4
|
POU domain, class 3, transcription factor 4 |
| chr4_+_44756608 | 0.62 |
ENSMUST00000143385.2
|
Zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr12_+_76593799 | 0.61 |
ENSMUST00000218380.2
ENSMUST00000219751.2 |
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr17_-_45125468 | 0.61 |
ENSMUST00000159943.8
ENSMUST00000160673.8 |
Runx2
|
runt related transcription factor 2 |
| chr13_+_47347301 | 0.60 |
ENSMUST00000110111.4
|
Rnf144b
|
ring finger protein 144B |
| chr7_+_127845984 | 0.59 |
ENSMUST00000164710.8
ENSMUST00000070656.12 |
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr2_-_45000389 | 0.59 |
ENSMUST00000201804.4
ENSMUST00000028229.13 ENSMUST00000202187.4 ENSMUST00000153561.6 ENSMUST00000201490.2 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr8_-_116434517 | 0.57 |
ENSMUST00000109104.2
|
Maf
|
avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr16_-_76169902 | 0.54 |
ENSMUST00000054178.8
|
Nrip1
|
nuclear receptor interacting protein 1 |
| chr15_-_76084035 | 0.46 |
ENSMUST00000054449.14
ENSMUST00000169714.8 ENSMUST00000165453.8 |
Plec
|
plectin |
| chrX_+_135723420 | 0.44 |
ENSMUST00000033800.13
|
Plp1
|
proteolipid protein (myelin) 1 |
| chr2_+_61423469 | 0.43 |
ENSMUST00000112494.2
|
Tank
|
TRAF family member-associated Nf-kappa B activator |
| chr4_+_44756553 | 0.42 |
ENSMUST00000107824.9
|
Zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr14_-_78774201 | 0.42 |
ENSMUST00000123853.9
|
Akap11
|
A kinase (PRKA) anchor protein 11 |
| chr1_+_135075377 | 0.41 |
ENSMUST00000125774.2
|
Arl8a
|
ADP-ribosylation factor-like 8A |
| chr9_+_21077010 | 0.38 |
ENSMUST00000039413.15
|
Pde4a
|
phosphodiesterase 4A, cAMP specific |
| chr6_-_37419030 | 0.37 |
ENSMUST00000041093.6
|
Creb3l2
|
cAMP responsive element binding protein 3-like 2 |
| chr7_-_127837154 | 0.35 |
ENSMUST00000078816.5
|
9130023H24Rik
|
RIKEN cDNA 9130023H24 gene |
| chr1_-_87322443 | 0.35 |
ENSMUST00000113212.4
|
Kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chrX_-_72868544 | 0.34 |
ENSMUST00000002080.12
ENSMUST00000114438.3 |
Pdzd4
|
PDZ domain containing 4 |
| chr11_+_66802807 | 0.33 |
ENSMUST00000123434.3
|
Pirt
|
phosphoinositide-interacting regulator of transient receptor potential channels |
| chr10_+_128061699 | 0.33 |
ENSMUST00000026455.8
|
Mip
|
major intrinsic protein of lens fiber |
| chr2_-_45001141 | 0.31 |
ENSMUST00000201969.4
ENSMUST00000201623.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr2_+_61423421 | 0.31 |
ENSMUST00000112495.8
ENSMUST00000112501.9 |
Tank
|
TRAF family member-associated Nf-kappa B activator |
| chrX_+_158242121 | 0.31 |
ENSMUST00000112470.3
ENSMUST00000043151.12 ENSMUST00000156172.3 |
Map7d2
|
MAP7 domain containing 2 |
| chr9_+_107458495 | 0.30 |
ENSMUST00000040059.9
|
Hyal3
|
hyaluronoglucosaminidase 3 |
| chr10_+_53213763 | 0.30 |
ENSMUST00000219491.2
ENSMUST00000163319.9 ENSMUST00000220197.2 ENSMUST00000046221.8 ENSMUST00000218468.2 ENSMUST00000219921.2 |
Pln
|
phospholamban |
| chr5_+_139408906 | 0.30 |
ENSMUST00000066211.5
|
Gper1
|
G protein-coupled estrogen receptor 1 |
| chr10_-_81436671 | 0.29 |
ENSMUST00000151858.8
ENSMUST00000142948.8 ENSMUST00000072020.9 |
Tle6
|
transducin-like enhancer of split 6 |
| chr7_+_127846121 | 0.29 |
ENSMUST00000167965.8
|
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr15_-_64254754 | 0.29 |
ENSMUST00000177374.8
ENSMUST00000110114.10 ENSMUST00000110115.9 ENSMUST00000023008.16 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr19_+_23881821 | 0.29 |
ENSMUST00000237688.2
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr11_-_94133527 | 0.28 |
ENSMUST00000061469.4
|
Wfikkn2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr5_-_53864595 | 0.27 |
ENSMUST00000200691.4
|
Cckar
|
cholecystokinin A receptor |
| chr7_+_4693603 | 0.24 |
ENSMUST00000120836.8
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr8_+_91681550 | 0.24 |
ENSMUST00000210947.2
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr7_-_119461027 | 0.24 |
ENSMUST00000137888.2
ENSMUST00000142120.2 |
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr5_-_53864874 | 0.23 |
ENSMUST00000031093.5
|
Cckar
|
cholecystokinin A receptor |
| chr6_-_113478779 | 0.21 |
ENSMUST00000101059.4
ENSMUST00000204268.3 ENSMUST00000205170.2 ENSMUST00000205075.2 ENSMUST00000204134.3 |
Prrt3
|
proline-rich transmembrane protein 3 |
| chr7_-_4781140 | 0.21 |
ENSMUST00000094892.12
|
Il11
|
interleukin 11 |
| chr1_+_183766572 | 0.21 |
ENSMUST00000048655.8
|
Dusp10
|
dual specificity phosphatase 10 |
| chr7_+_4693759 | 0.20 |
ENSMUST00000048248.9
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr16_+_32151056 | 0.20 |
ENSMUST00000115151.5
ENSMUST00000232137.2 |
Ubxn7
|
UBX domain protein 7 |
| chr9_-_32454157 | 0.20 |
ENSMUST00000183767.2
|
Fli1
|
Friend leukemia integration 1 |
| chr11_+_96820091 | 0.20 |
ENSMUST00000054311.6
ENSMUST00000107636.4 |
Prr15l
|
proline rich 15-like |
| chr1_+_86230931 | 0.19 |
ENSMUST00000113306.4
|
B3gnt7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr14_-_70864448 | 0.19 |
ENSMUST00000110984.4
|
Dmtn
|
dematin actin binding protein |
| chr1_+_60448931 | 0.18 |
ENSMUST00000189082.7
ENSMUST00000187709.7 |
Abi2
|
abl interactor 2 |
| chr16_+_44913974 | 0.18 |
ENSMUST00000099498.10
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr16_+_44914397 | 0.17 |
ENSMUST00000061050.6
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr17_+_44445659 | 0.17 |
ENSMUST00000239215.2
|
Clic5
|
chloride intracellular channel 5 |
| chr2_+_153187729 | 0.16 |
ENSMUST00000227428.2
ENSMUST00000109790.2 |
Asxl1
|
ASXL transcriptional regulator 1 |
| chr11_-_95201012 | 0.15 |
ENSMUST00000103159.10
ENSMUST00000107734.10 ENSMUST00000107733.10 |
Kat7
|
K(lysine) acetyltransferase 7 |
| chr19_+_46385448 | 0.15 |
ENSMUST00000118440.3
|
Sufu
|
SUFU negative regulator of hedgehog signaling |
| chr18_-_60881679 | 0.14 |
ENSMUST00000237783.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr2_-_32977182 | 0.14 |
ENSMUST00000102810.10
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr15_-_64254585 | 0.14 |
ENSMUST00000176384.8
ENSMUST00000175799.8 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr17_-_45125537 | 0.13 |
ENSMUST00000113571.10
|
Runx2
|
runt related transcription factor 2 |
| chr8_-_3722290 | 0.13 |
ENSMUST00000159364.3
|
Fcor
|
Foxo1 corepressor |
| chr16_-_36486429 | 0.12 |
ENSMUST00000089620.11
|
Cd86
|
CD86 antigen |
| chr6_+_124973752 | 0.12 |
ENSMUST00000162000.4
|
Pianp
|
PILR alpha associated neural protein |
| chr5_+_3391477 | 0.12 |
ENSMUST00000199156.2
|
Cdk6
|
cyclin-dependent kinase 6 |
| chr3_+_58913234 | 0.11 |
ENSMUST00000040846.15
|
Med12l
|
mediator complex subunit 12-like |
| chr2_-_90900628 | 0.11 |
ENSMUST00000111436.3
ENSMUST00000073575.12 |
Slc39a13
|
solute carrier family 39 (metal ion transporter), member 13 |
| chr14_-_70864666 | 0.10 |
ENSMUST00000022694.17
|
Dmtn
|
dematin actin binding protein |
| chr1_+_60448813 | 0.10 |
ENSMUST00000188594.7
ENSMUST00000188618.7 ENSMUST00000189980.7 |
Abi2
|
abl interactor 2 |
| chr1_+_153541020 | 0.10 |
ENSMUST00000152114.8
ENSMUST00000111812.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr2_-_125565307 | 0.10 |
ENSMUST00000042246.14
|
Shc4
|
SHC (Src homology 2 domain containing) family, member 4 |
| chr4_+_109200225 | 0.09 |
ENSMUST00000030281.12
|
Eps15
|
epidermal growth factor receptor pathway substrate 15 |
| chr10_-_128334515 | 0.09 |
ENSMUST00000026428.4
|
Myl6b
|
myosin, light polypeptide 6B |
| chr1_+_60448703 | 0.09 |
ENSMUST00000052332.15
|
Abi2
|
abl interactor 2 |
| chr19_+_37674029 | 0.08 |
ENSMUST00000073391.5
|
Cyp26c1
|
cytochrome P450, family 26, subfamily c, polypeptide 1 |
| chr10_+_7668655 | 0.08 |
ENSMUST00000015901.11
|
Ppil4
|
peptidylprolyl isomerase (cyclophilin)-like 4 |
| chr4_+_128548479 | 0.08 |
ENSMUST00000030588.13
ENSMUST00000136377.8 |
Phc2
|
polyhomeotic 2 |
| chr1_-_183766195 | 0.07 |
ENSMUST00000050306.8
|
1700056E22Rik
|
RIKEN cDNA 1700056E22 gene |
| chr8_-_47742389 | 0.07 |
ENSMUST00000211737.2
|
Stox2
|
storkhead box 2 |
| chr1_+_86231208 | 0.07 |
ENSMUST00000188695.2
|
B3gnt7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chrX_+_135723531 | 0.06 |
ENSMUST00000113085.2
|
Plp1
|
proteolipid protein (myelin) 1 |
| chr12_-_84923252 | 0.06 |
ENSMUST00000163189.8
ENSMUST00000110254.9 ENSMUST00000002073.13 |
Ltbp2
|
latent transforming growth factor beta binding protein 2 |
| chr6_+_124973644 | 0.06 |
ENSMUST00000032479.11
|
Pianp
|
PILR alpha associated neural protein |
| chr2_+_3119558 | 0.06 |
ENSMUST00000062934.7
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr2_+_3119442 | 0.06 |
ENSMUST00000091505.11
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr2_+_3119371 | 0.06 |
ENSMUST00000115099.9
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr2_-_79287095 | 0.06 |
ENSMUST00000041099.5
|
Neurod1
|
neurogenic differentiation 1 |
| chr16_-_29360301 | 0.06 |
ENSMUST00000057018.15
ENSMUST00000182627.8 |
Atp13a4
|
ATPase type 13A4 |
| chr18_+_65715460 | 0.05 |
ENSMUST00000169679.8
ENSMUST00000183326.2 |
Zfp532
|
zinc finger protein 532 |
| chr12_+_55646209 | 0.05 |
ENSMUST00000051857.5
|
Insm2
|
insulinoma-associated 2 |
| chr18_+_36431732 | 0.04 |
ENSMUST00000210490.3
|
Igip
|
IgA inducing protein |
| chr15_-_81742686 | 0.04 |
ENSMUST00000050467.9
ENSMUST00000231000.2 |
Tob2
|
transducer of ERBB2, 2 |
| chr9_-_72019053 | 0.04 |
ENSMUST00000183492.8
ENSMUST00000184523.8 ENSMUST00000034755.13 |
Tcf12
|
transcription factor 12 |
| chr7_-_127214372 | 0.04 |
ENSMUST00000128731.8
ENSMUST00000122066.8 |
Zfp629
|
zinc finger protein 629 |
| chr3_-_79053182 | 0.04 |
ENSMUST00000118340.7
|
Rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr1_+_24234268 | 0.04 |
ENSMUST00000088349.3
|
Col9a1
|
collagen, type IX, alpha 1 |
| chr8_+_91681898 | 0.03 |
ENSMUST00000209746.2
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr17_+_25097199 | 0.03 |
ENSMUST00000050714.8
|
Igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr19_+_8595369 | 0.02 |
ENSMUST00000010250.4
|
Slc22a6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr11_+_69920542 | 0.02 |
ENSMUST00000232266.2
ENSMUST00000132597.5 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr15_-_79897404 | 0.02 |
ENSMUST00000229912.2
ENSMUST00000229795.2 |
Pdgfb
|
platelet derived growth factor, B polypeptide |
| chrX_+_142936691 | 0.02 |
ENSMUST00000135687.2
|
A730046J19Rik
|
RIKEN cDNA A730046J19 gene |
| chr5_-_41921834 | 0.02 |
ENSMUST00000060820.8
|
Nkx3-2
|
NK3 homeobox 2 |
| chrX_-_74423647 | 0.02 |
ENSMUST00000114085.9
|
F8
|
coagulation factor VIII |
| chr10_+_75425171 | 0.01 |
ENSMUST00000072217.9
|
Ggt5
|
gamma-glutamyltransferase 5 |
| chr2_+_31649946 | 0.01 |
ENSMUST00000028190.13
|
Abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr19_+_46385321 | 0.01 |
ENSMUST00000039922.13
ENSMUST00000111867.9 ENSMUST00000120778.8 |
Sufu
|
SUFU negative regulator of hedgehog signaling |
| chr4_-_133981387 | 0.01 |
ENSMUST00000060050.6
|
Grrp1
|
glycine/arginine rich protein 1 |
| chr1_-_154692678 | 0.01 |
ENSMUST00000238369.2
|
Cacna1e
|
calcium channel, voltage-dependent, R type, alpha 1E subunit |
| chr18_+_23886765 | 0.01 |
ENSMUST00000115830.8
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr11_-_102710490 | 0.01 |
ENSMUST00000092567.11
|
Gjc1
|
gap junction protein, gamma 1 |
| chr7_+_18817767 | 0.01 |
ENSMUST00000032568.14
ENSMUST00000122999.8 ENSMUST00000108473.10 ENSMUST00000108474.2 ENSMUST00000238982.2 |
Dmpk
|
dystrophia myotonica-protein kinase |
| chrY_+_17400761 | 0.00 |
ENSMUST00000179408.3
|
Rbm31y
|
RNA binding motif 31, Y-linked |
| chr12_-_109019507 | 0.00 |
ENSMUST00000185745.2
ENSMUST00000239108.2 |
Begain
|
brain-enriched guanylate kinase-associated |
| chr17_+_23819818 | 0.00 |
ENSMUST00000095595.9
ENSMUST00000115509.8 ENSMUST00000120967.8 ENSMUST00000148062.8 ENSMUST00000129227.8 |
Zscan10
|
zinc finger and SCAN domain containing 10 |
| chr12_-_17226889 | 0.00 |
ENSMUST00000170580.3
|
Kcnf1
|
potassium voltage-gated channel, subfamily F, member 1 |
| chr8_-_49008305 | 0.00 |
ENSMUST00000110346.9
ENSMUST00000211976.2 |
Tenm3
|
teneurin transmembrane protein 3 |
| chr9_-_65424281 | 0.00 |
ENSMUST00000061766.6
|
Ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr10_-_95253042 | 0.00 |
ENSMUST00000135822.8
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr2_-_179867605 | 0.00 |
ENSMUST00000015791.6
|
Lama5
|
laminin, alpha 5 |
| chr16_+_29884153 | 0.00 |
ENSMUST00000023171.8
|
Hes1
|
hes family bHLH transcription factor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp652
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.3 | 1.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.3 | 0.8 | GO:1904956 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.2 | 0.7 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.2 | 0.7 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.1 | 0.5 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 0.9 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.3 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 0.1 | 0.7 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.9 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.5 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) positive regulation of somatostatin secretion(GO:0090274) |
| 0.1 | 0.7 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.4 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 0.2 | GO:0072708 | response to sorbitol(GO:0072708) response to dithiothreitol(GO:0072720) |
| 0.0 | 0.3 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.0 | 0.3 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.2 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:0036275 | response to 5-fluorouracil(GO:0036275) |
| 0.0 | 0.6 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 | 0.4 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.2 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.2 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.3 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.0 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.0 | 0.5 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.8 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.6 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 1.0 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.3 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.0 | GO:0072244 | metanephric glomerular epithelium development(GO:0072244) |
| 0.0 | 0.2 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.0 | 0.2 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.1 | GO:0002911 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.3 | GO:0048266 | behavioral response to pain(GO:0048266) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.3 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0097132 | cyclin D2-CDK6 complex(GO:0097132) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.0 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 0.7 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.9 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.3 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.1 | 0.7 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 0.8 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 1.1 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.2 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.4 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.4 | GO:0030552 | cAMP binding(GO:0030552) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |