Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
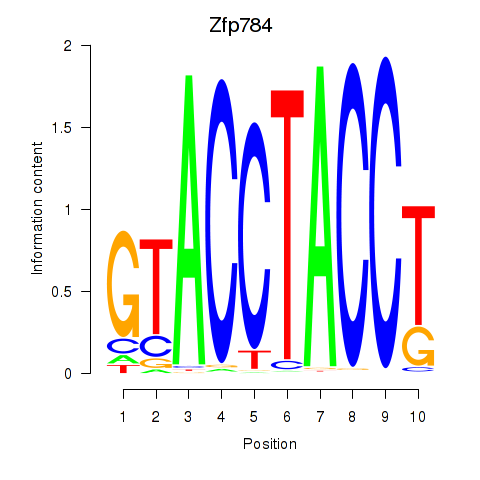
Results for Zfp784
Z-value: 0.62
Transcription factors associated with Zfp784
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp784
|
ENSMUSG00000043290.7 | zinc finger protein 784 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp784 | mm39_v1_chr7_-_5041427_5041450 | 0.81 | 9.3e-02 | Click! |
Activity profile of Zfp784 motif
Sorted Z-values of Zfp784 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_82045800 | 0.47 |
ENSMUST00000235015.2
ENSMUST00000163123.3 |
Slc8a1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr6_-_28831746 | 0.45 |
ENSMUST00000062304.7
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr13_+_21901791 | 0.39 |
ENSMUST00000188775.2
|
H3c10
|
H3 clustered histone 10 |
| chr13_+_47347301 | 0.37 |
ENSMUST00000110111.4
|
Rnf144b
|
ring finger protein 144B |
| chr5_+_64969679 | 0.33 |
ENSMUST00000166409.6
ENSMUST00000197879.2 |
Klf3
|
Kruppel-like factor 3 (basic) |
| chr2_+_101455022 | 0.32 |
ENSMUST00000044031.4
|
Rag2
|
recombination activating gene 2 |
| chr1_+_151447232 | 0.30 |
ENSMUST00000111875.2
|
Niban1
|
niban apoptosis regulator 1 |
| chr19_+_8966641 | 0.28 |
ENSMUST00000092956.4
ENSMUST00000092955.11 |
Ahnak
|
AHNAK nucleoprotein (desmoyokin) |
| chr9_-_119897358 | 0.27 |
ENSMUST00000064165.5
|
Cx3cr1
|
chemokine (C-X3-C motif) receptor 1 |
| chr17_-_38618461 | 0.27 |
ENSMUST00000213505.2
|
Olfr137
|
olfactory receptor 137 |
| chr9_-_119897328 | 0.26 |
ENSMUST00000177637.2
|
Cx3cr1
|
chemokine (C-X3-C motif) receptor 1 |
| chr1_+_151447124 | 0.25 |
ENSMUST00000148810.8
|
Niban1
|
niban apoptosis regulator 1 |
| chr1_-_173569301 | 0.25 |
ENSMUST00000042610.15
ENSMUST00000127730.2 |
Ifi207
|
interferon activated gene 207 |
| chr11_+_50492899 | 0.25 |
ENSMUST00000142118.3
ENSMUST00000040523.9 |
Adamts2
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 2 |
| chrX_-_107877909 | 0.25 |
ENSMUST00000101283.4
ENSMUST00000150434.8 |
Brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr11_-_101917745 | 0.24 |
ENSMUST00000107167.2
ENSMUST00000062801.11 |
Mpp3
|
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) |
| chr15_+_102885467 | 0.24 |
ENSMUST00000001706.7
|
Hoxc9
|
homeobox C9 |
| chr17_+_6047112 | 0.23 |
ENSMUST00000115786.8
|
Synj2
|
synaptojanin 2 |
| chr12_+_76593799 | 0.23 |
ENSMUST00000218380.2
ENSMUST00000219751.2 |
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr16_-_3821614 | 0.22 |
ENSMUST00000171658.2
ENSMUST00000171762.2 |
Slx4
|
SLX4 structure-specific endonuclease subunit homolog (S. cerevisiae) |
| chr17_+_12338161 | 0.22 |
ENSMUST00000024594.9
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr11_+_118319029 | 0.22 |
ENSMUST00000124861.2
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr7_-_48530777 | 0.22 |
ENSMUST00000058745.15
|
E2f8
|
E2F transcription factor 8 |
| chr1_-_160862364 | 0.22 |
ENSMUST00000177003.2
ENSMUST00000159250.9 ENSMUST00000162226.9 |
Zbtb37
|
zinc finger and BTB domain containing 37 |
| chr16_-_19019100 | 0.21 |
ENSMUST00000103749.3
|
Iglc2
|
immunoglobulin lambda constant 2 |
| chr7_+_123582021 | 0.21 |
ENSMUST00000106437.2
|
Hs3st4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4 |
| chr10_-_30679289 | 0.21 |
ENSMUST00000215725.2
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr1_+_138891155 | 0.20 |
ENSMUST00000200533.5
|
Dennd1b
|
DENN/MADD domain containing 1B |
| chr11_+_70453666 | 0.20 |
ENSMUST00000072237.13
ENSMUST00000072873.14 |
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr11_+_70453724 | 0.19 |
ENSMUST00000102559.11
|
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr14_-_19261196 | 0.19 |
ENSMUST00000112797.12
ENSMUST00000225885.2 |
D830030K20Rik
|
RIKEN cDNA D830030K20 gene |
| chr16_-_16345197 | 0.19 |
ENSMUST00000069284.14
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr8_-_84996976 | 0.19 |
ENSMUST00000005120.12
ENSMUST00000163993.2 ENSMUST00000098578.10 |
Ccdc130
|
coiled-coil domain containing 130 |
| chr15_-_76082346 | 0.19 |
ENSMUST00000072692.11
|
Plec
|
plectin |
| chr6_+_29526679 | 0.18 |
ENSMUST00000167252.5
|
Irf5
|
interferon regulatory factor 5 |
| chr17_-_34340918 | 0.18 |
ENSMUST00000151986.2
|
Brd2
|
bromodomain containing 2 |
| chr1_+_92545510 | 0.18 |
ENSMUST00000213247.2
|
Olfr12
|
olfactory receptor 12 |
| chr4_+_152160713 | 0.17 |
ENSMUST00000239025.2
|
Plekhg5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr11_-_103158190 | 0.17 |
ENSMUST00000021324.3
|
Map3k14
|
mitogen-activated protein kinase kinase kinase 14 |
| chr13_-_38221045 | 0.17 |
ENSMUST00000089840.5
|
Cage1
|
cancer antigen 1 |
| chr18_+_34380738 | 0.17 |
ENSMUST00000066133.7
|
Apc
|
APC, WNT signaling pathway regulator |
| chr6_+_72324281 | 0.17 |
ENSMUST00000077783.5
|
0610030E20Rik
|
RIKEN cDNA 0610030E20 gene |
| chr19_-_12100560 | 0.16 |
ENSMUST00000213471.2
ENSMUST00000214676.2 |
Olfr76
|
olfactory receptor 76 |
| chr15_-_50753437 | 0.16 |
ENSMUST00000077935.6
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr9_-_87137515 | 0.16 |
ENSMUST00000093802.6
|
Cep162
|
centrosomal protein 162 |
| chr3_-_136031799 | 0.15 |
ENSMUST00000196159.5
ENSMUST00000041577.13 |
Bank1
|
B cell scaffold protein with ankyrin repeats 1 |
| chr15_+_80861966 | 0.15 |
ENSMUST00000139517.9
ENSMUST00000137255.3 ENSMUST00000137004.2 |
Sgsm3
|
small G protein signaling modulator 3 |
| chr16_-_97883632 | 0.15 |
ENSMUST00000231268.2
ENSMUST00000231741.2 |
A630089N07Rik
|
RIKEN cDNA A630089N07 gene |
| chr9_-_97915036 | 0.15 |
ENSMUST00000162295.2
|
Clstn2
|
calsyntenin 2 |
| chr16_-_31133622 | 0.14 |
ENSMUST00000115230.2
ENSMUST00000130560.8 |
Apod
|
apolipoprotein D |
| chr2_+_111120481 | 0.14 |
ENSMUST00000215210.2
ENSMUST00000216229.2 |
Olfr1278
|
olfactory receptor 1278 |
| chr8_-_43759973 | 0.14 |
ENSMUST00000211248.2
ENSMUST00000209356.2 |
Zfp42
|
zinc finger protein 42 |
| chr6_+_29526624 | 0.14 |
ENSMUST00000004392.12
|
Irf5
|
interferon regulatory factor 5 |
| chr7_+_142025817 | 0.14 |
ENSMUST00000105966.2
|
Lsp1
|
lymphocyte specific 1 |
| chr19_+_18609343 | 0.13 |
ENSMUST00000159572.9
|
Nmrk1
|
nicotinamide riboside kinase 1 |
| chr5_-_137609691 | 0.13 |
ENSMUST00000031731.14
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr5_-_123887434 | 0.12 |
ENSMUST00000182955.8
ENSMUST00000182489.8 ENSMUST00000183147.9 ENSMUST00000050827.14 ENSMUST00000057795.12 ENSMUST00000111515.8 ENSMUST00000182309.8 |
Rsrc2
|
arginine/serine-rich coiled-coil 2 |
| chr7_+_101583283 | 0.12 |
ENSMUST00000209639.2
ENSMUST00000210679.2 |
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr11_+_68393845 | 0.12 |
ENSMUST00000102613.8
ENSMUST00000060441.7 |
Pik3r6
|
phosphoinositide-3-kinase regulatory subunit 5 |
| chr19_+_18609291 | 0.12 |
ENSMUST00000042392.14
ENSMUST00000237347.2 |
Nmrk1
|
nicotinamide riboside kinase 1 |
| chr17_+_44499451 | 0.12 |
ENSMUST00000024755.7
|
Clic5
|
chloride intracellular channel 5 |
| chr15_+_102898966 | 0.12 |
ENSMUST00000001703.8
|
Hoxc8
|
homeobox C8 |
| chr9_+_110074574 | 0.12 |
ENSMUST00000197850.5
|
Cspg5
|
chondroitin sulfate proteoglycan 5 |
| chr7_-_103463120 | 0.12 |
ENSMUST00000098192.4
|
Hbb-bt
|
hemoglobin, beta adult t chain |
| chr11_+_97206542 | 0.12 |
ENSMUST00000019026.10
ENSMUST00000132168.2 |
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr11_+_75542328 | 0.11 |
ENSMUST00000069057.13
|
Myo1c
|
myosin IC |
| chr1_+_40363701 | 0.11 |
ENSMUST00000095020.9
ENSMUST00000194296.6 |
Il1rl2
|
interleukin 1 receptor-like 2 |
| chr14_-_25769457 | 0.11 |
ENSMUST00000069180.8
|
Zcchc24
|
zinc finger, CCHC domain containing 24 |
| chr13_-_64422775 | 0.11 |
ENSMUST00000221634.2
ENSMUST00000039318.16 |
Cdc14b
|
CDC14 cell division cycle 14B |
| chr17_-_74017410 | 0.11 |
ENSMUST00000112591.3
ENSMUST00000024858.12 |
Galnt14
|
polypeptide N-acetylgalactosaminyltransferase 14 |
| chr19_+_23881821 | 0.11 |
ENSMUST00000237688.2
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr17_-_56428968 | 0.11 |
ENSMUST00000041357.9
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr6_-_124519240 | 0.10 |
ENSMUST00000159463.8
ENSMUST00000162844.2 ENSMUST00000160505.8 ENSMUST00000162443.8 |
C1s1
|
complement component 1, s subcomponent 1 |
| chr14_-_70864448 | 0.10 |
ENSMUST00000110984.4
|
Dmtn
|
dematin actin binding protein |
| chr7_+_142025575 | 0.10 |
ENSMUST00000038946.9
|
Lsp1
|
lymphocyte specific 1 |
| chr7_+_4925781 | 0.10 |
ENSMUST00000207527.2
ENSMUST00000207687.2 ENSMUST00000208754.2 |
Nat14
|
N-acetyltransferase 14 |
| chr5_-_137609634 | 0.10 |
ENSMUST00000054564.13
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr13_-_67569483 | 0.10 |
ENSMUST00000224113.2
ENSMUST00000223644.2 ENSMUST00000056470.10 |
Zfp459
|
zinc finger protein 459 |
| chr7_-_6511472 | 0.10 |
ENSMUST00000207055.3
ENSMUST00000209097.2 ENSMUST00000208623.3 ENSMUST00000208207.2 ENSMUST00000209029.4 ENSMUST00000214383.2 ENSMUST00000207624.2 ENSMUST00000213549.2 |
Olfr1348
|
olfactory receptor 1348 |
| chr6_+_31375670 | 0.10 |
ENSMUST00000026699.15
|
Mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr10_-_30679071 | 0.10 |
ENSMUST00000068567.5
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chrX_-_149372840 | 0.10 |
ENSMUST00000112725.8
ENSMUST00000112720.8 |
Apex2
|
apurinic/apyrimidinic endonuclease 2 |
| chr6_-_118539187 | 0.09 |
ENSMUST00000112830.3
|
Ankrd26
|
ankyrin repeat domain 26 |
| chr2_+_146063841 | 0.09 |
ENSMUST00000089257.6
|
Insm1
|
insulinoma-associated 1 |
| chr11_+_70453806 | 0.09 |
ENSMUST00000079244.12
ENSMUST00000102558.11 |
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr6_-_99412306 | 0.09 |
ENSMUST00000113322.9
ENSMUST00000176850.8 ENSMUST00000176632.8 |
Foxp1
|
forkhead box P1 |
| chr19_-_12093187 | 0.09 |
ENSMUST00000208391.3
ENSMUST00000214103.2 |
Olfr1428
|
olfactory receptor 1428 |
| chr11_+_75542902 | 0.09 |
ENSMUST00000102504.10
|
Myo1c
|
myosin IC |
| chr15_+_76544763 | 0.08 |
ENSMUST00000004294.12
|
Kifc2
|
kinesin family member C2 |
| chr13_+_14238361 | 0.08 |
ENSMUST00000129488.8
ENSMUST00000110536.8 ENSMUST00000110534.8 ENSMUST00000039538.15 ENSMUST00000110533.2 |
Arid4b
|
AT rich interactive domain 4B (RBP1-like) |
| chr5_-_53864595 | 0.08 |
ENSMUST00000200691.4
|
Cckar
|
cholecystokinin A receptor |
| chr11_+_107438751 | 0.08 |
ENSMUST00000100305.8
ENSMUST00000075012.8 ENSMUST00000106746.8 |
Helz
|
helicase with zinc finger domain |
| chr5_-_53864874 | 0.08 |
ENSMUST00000031093.5
|
Cckar
|
cholecystokinin A receptor |
| chr7_-_105230807 | 0.08 |
ENSMUST00000191011.7
|
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr2_+_18069375 | 0.08 |
ENSMUST00000114671.8
ENSMUST00000114680.9 |
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr2_+_111355548 | 0.08 |
ENSMUST00000217845.3
|
Olfr1293-ps
|
olfactory receptor 1293, pseudogene |
| chr5_+_30745447 | 0.08 |
ENSMUST00000066295.5
|
Kcnk3
|
potassium channel, subfamily K, member 3 |
| chr11_+_5049608 | 0.08 |
ENSMUST00000139742.8
|
Rhbdd3
|
rhomboid domain containing 3 |
| chr11_+_92989229 | 0.08 |
ENSMUST00000107859.8
ENSMUST00000107861.8 ENSMUST00000042943.13 ENSMUST00000107858.9 |
Car10
|
carbonic anhydrase 10 |
| chr7_+_30215124 | 0.08 |
ENSMUST00000238768.2
|
Arhgap33os
|
Rho GTPase activating protein 33, opposite strand |
| chr15_-_78456898 | 0.08 |
ENSMUST00000043214.8
|
Rac2
|
Rac family small GTPase 2 |
| chr1_-_133681419 | 0.08 |
ENSMUST00000125659.8
ENSMUST00000048953.14 ENSMUST00000165602.9 |
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr11_+_73051228 | 0.08 |
ENSMUST00000006104.10
ENSMUST00000135202.8 ENSMUST00000136894.3 |
P2rx5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr17_-_47748289 | 0.08 |
ENSMUST00000061885.9
|
1700001C19Rik
|
RIKEN cDNA 1700001C19 gene |
| chr3_-_136031723 | 0.08 |
ENSMUST00000198206.2
|
Bank1
|
B cell scaffold protein with ankyrin repeats 1 |
| chr9_-_50639230 | 0.08 |
ENSMUST00000118707.2
ENSMUST00000034566.15 |
Dixdc1
|
DIX domain containing 1 |
| chr3_-_108117754 | 0.08 |
ENSMUST00000117784.8
ENSMUST00000119650.8 ENSMUST00000117409.8 |
Atxn7l2
|
ataxin 7-like 2 |
| chr4_+_135639117 | 0.07 |
ENSMUST00000068830.4
|
Cnr2
|
cannabinoid receptor 2 (macrophage) |
| chr2_-_45003270 | 0.07 |
ENSMUST00000202935.4
ENSMUST00000068415.11 ENSMUST00000127520.8 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr3_+_125474628 | 0.07 |
ENSMUST00000174648.6
|
Ndst4
|
N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 |
| chrX_+_150961960 | 0.07 |
ENSMUST00000096275.5
|
Iqsec2
|
IQ motif and Sec7 domain 2 |
| chr2_+_72115981 | 0.07 |
ENSMUST00000090824.12
ENSMUST00000135469.8 |
Map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr10_+_7556948 | 0.07 |
ENSMUST00000165952.9
|
Lats1
|
large tumor suppressor |
| chr2_-_9883391 | 0.07 |
ENSMUST00000102976.4
|
Gata3
|
GATA binding protein 3 |
| chr14_+_21549798 | 0.07 |
ENSMUST00000182855.8
ENSMUST00000182405.9 ENSMUST00000069648.14 |
Kat6b
|
K(lysine) acetyltransferase 6B |
| chr7_-_105230479 | 0.07 |
ENSMUST00000191601.7
|
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr6_-_124710030 | 0.06 |
ENSMUST00000173647.2
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr9_+_44151962 | 0.06 |
ENSMUST00000092426.5
ENSMUST00000217221.2 ENSMUST00000213891.2 |
Ccdc153
|
coiled-coil domain containing 153 |
| chr16_-_16962279 | 0.06 |
ENSMUST00000232033.2
ENSMUST00000231597.2 ENSMUST00000232540.2 |
Ccdc116
|
coiled-coil domain containing 116 |
| chr2_-_45002902 | 0.06 |
ENSMUST00000076836.13
ENSMUST00000176732.8 ENSMUST00000200844.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr7_+_25318829 | 0.06 |
ENSMUST00000077338.12
ENSMUST00000085953.5 |
Dmac2
|
distal membrane arm assembly complex 2 |
| chr19_+_29344846 | 0.06 |
ENSMUST00000016640.8
|
Cd274
|
CD274 antigen |
| chr7_-_105230698 | 0.06 |
ENSMUST00000189378.7
|
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr7_-_104577264 | 0.06 |
ENSMUST00000215359.2
|
Olfr668
|
olfactory receptor 668 |
| chr14_-_70864666 | 0.06 |
ENSMUST00000022694.17
|
Dmtn
|
dematin actin binding protein |
| chr15_-_50753792 | 0.06 |
ENSMUST00000185183.2
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr11_-_34674677 | 0.06 |
ENSMUST00000093193.12
ENSMUST00000101365.9 |
Dock2
|
dedicator of cyto-kinesis 2 |
| chr6_+_29361408 | 0.06 |
ENSMUST00000156163.2
|
Calu
|
calumenin |
| chr7_-_24705320 | 0.06 |
ENSMUST00000102858.10
ENSMUST00000196684.2 ENSMUST00000080882.11 |
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr4_+_62537750 | 0.06 |
ENSMUST00000084521.11
ENSMUST00000107424.2 |
Rgs3
|
regulator of G-protein signaling 3 |
| chr18_-_10610332 | 0.05 |
ENSMUST00000025142.13
|
Esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr10_-_80895949 | 0.05 |
ENSMUST00000219401.2
ENSMUST00000220317.2 ENSMUST00000005067.6 ENSMUST00000218208.2 |
Sgta
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr4_-_152561896 | 0.05 |
ENSMUST00000238738.2
ENSMUST00000162017.3 ENSMUST00000030768.10 |
Kcnab2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr4_+_62327010 | 0.05 |
ENSMUST00000084524.4
|
Prpf4
|
pre-mRNA processing factor 4 |
| chr2_-_23462052 | 0.05 |
ENSMUST00000132484.7
|
Spopl
|
speckle-type BTB/POZ protein-like |
| chr5_-_93192881 | 0.05 |
ENSMUST00000061328.6
|
Sowahb
|
sosondowah ankyrin repeat domain family member B |
| chr2_+_70392351 | 0.05 |
ENSMUST00000094934.11
|
Gad1
|
glutamate decarboxylase 1 |
| chr14_-_65662609 | 0.05 |
ENSMUST00000224594.2
|
Pnoc
|
prepronociceptin |
| chr15_+_32244947 | 0.05 |
ENSMUST00000067458.7
|
Sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr3_+_90561560 | 0.05 |
ENSMUST00000079286.4
|
S100a7a
|
S100 calcium binding protein A7A |
| chr9_-_48876290 | 0.05 |
ENSMUST00000008734.5
|
Htr3b
|
5-hydroxytryptamine (serotonin) receptor 3B |
| chr4_+_131649001 | 0.05 |
ENSMUST00000094666.4
|
Tmem200b
|
transmembrane protein 200B |
| chr8_+_45960804 | 0.05 |
ENSMUST00000067065.14
ENSMUST00000124544.8 ENSMUST00000138049.9 ENSMUST00000132139.9 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr14_-_65662902 | 0.05 |
ENSMUST00000059339.6
|
Pnoc
|
prepronociceptin |
| chr19_-_5944098 | 0.05 |
ENSMUST00000055911.6
ENSMUST00000236767.2 |
Tigd3
|
tigger transposable element derived 3 |
| chr10_-_100323122 | 0.05 |
ENSMUST00000128009.2
ENSMUST00000134477.2 ENSMUST00000099318.10 ENSMUST00000058154.15 |
Tmtc3
|
transmembrane and tetratricopeptide repeat containing 3 |
| chr18_+_34892599 | 0.05 |
ENSMUST00000097622.4
|
Fam53c
|
family with sequence similarity 53, member C |
| chr13_+_20978283 | 0.04 |
ENSMUST00000021757.5
ENSMUST00000221982.2 |
Aoah
|
acyloxyacyl hydrolase |
| chr9_+_108883907 | 0.04 |
ENSMUST00000154184.5
|
Shisa5
|
shisa family member 5 |
| chr6_+_15721086 | 0.04 |
ENSMUST00000120512.8
|
Mdfic
|
MyoD family inhibitor domain containing |
| chr13_-_64422693 | 0.04 |
ENSMUST00000109770.2
|
Cdc14b
|
CDC14 cell division cycle 14B |
| chrX_+_6489313 | 0.04 |
ENSMUST00000089520.3
|
Shroom4
|
shroom family member 4 |
| chr8_-_34237752 | 0.04 |
ENSMUST00000179364.3
|
Smim18
|
small integral membrane protein 18 |
| chr7_-_141017742 | 0.04 |
ENSMUST00000019226.14
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr2_-_163261439 | 0.04 |
ENSMUST00000046908.10
|
Oser1
|
oxidative stress responsive serine rich 1 |
| chr7_-_101582987 | 0.04 |
ENSMUST00000106964.8
ENSMUST00000106963.2 ENSMUST00000078448.11 ENSMUST00000106966.8 |
Lrrc51
|
leucine rich repeat containing 51 |
| chr8_+_105066924 | 0.04 |
ENSMUST00000212081.2
|
Cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr2_+_118219096 | 0.04 |
ENSMUST00000110877.8
ENSMUST00000005233.12 |
Eif2ak4
|
eukaryotic translation initiation factor 2 alpha kinase 4 |
| chr7_-_127529238 | 0.04 |
ENSMUST00000032988.10
ENSMUST00000206124.2 |
Prss8
|
protease, serine 8 (prostasin) |
| chr5_-_92190859 | 0.04 |
ENSMUST00000069937.11
ENSMUST00000086978.12 |
Cdkl2
|
cyclin-dependent kinase-like 2 (CDC2-related kinase) |
| chr17_-_48235560 | 0.04 |
ENSMUST00000113265.8
|
Foxp4
|
forkhead box P4 |
| chr8_+_105067159 | 0.03 |
ENSMUST00000212948.2
ENSMUST00000034343.5 |
Cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr11_+_94455865 | 0.03 |
ENSMUST00000040418.9
|
Chad
|
chondroadherin |
| chr17_-_48235325 | 0.03 |
ENSMUST00000113263.8
ENSMUST00000097311.9 |
Foxp4
|
forkhead box P4 |
| chr19_-_38807600 | 0.03 |
ENSMUST00000025963.8
|
Noc3l
|
NOC3 like DNA replication regulator |
| chr17_-_36582721 | 0.03 |
ENSMUST00000042717.13
|
Trim39
|
tripartite motif-containing 39 |
| chr14_-_63482668 | 0.03 |
ENSMUST00000118022.8
|
Gata4
|
GATA binding protein 4 |
| chr6_-_57668992 | 0.03 |
ENSMUST00000053386.6
|
Pyurf
|
Pigy upstream reading frame |
| chr11_+_66943453 | 0.03 |
ENSMUST00000108690.10
ENSMUST00000092996.5 |
Sco1
|
SCO1 cytochrome c oxidase assembly protein |
| chr14_-_63482720 | 0.03 |
ENSMUST00000067417.10
|
Gata4
|
GATA binding protein 4 |
| chr16_+_35590745 | 0.03 |
ENSMUST00000231579.2
|
Hspbap1
|
Hspb associated protein 1 |
| chr1_-_132470672 | 0.03 |
ENSMUST00000086521.11
|
Cntn2
|
contactin 2 |
| chr6_+_15720653 | 0.03 |
ENSMUST00000101663.10
ENSMUST00000190255.7 ENSMUST00000189359.7 ENSMUST00000125326.8 |
Mdfic
|
MyoD family inhibitor domain containing |
| chr17_+_43671314 | 0.03 |
ENSMUST00000226087.2
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr8_-_106363982 | 0.03 |
ENSMUST00000182863.2
ENSMUST00000194091.6 |
Gm5914
Agrp
|
predicted gene 5914 agouti related neuropeptide |
| chr1_-_82564501 | 0.03 |
ENSMUST00000087050.7
|
Col4a4
|
collagen, type IV, alpha 4 |
| chr1_-_36484328 | 0.02 |
ENSMUST00000125304.8
ENSMUST00000115011.8 |
Lman2l
|
lectin, mannose-binding 2-like |
| chr10_-_20600797 | 0.02 |
ENSMUST00000020165.14
|
Pde7b
|
phosphodiesterase 7B |
| chr7_-_73191484 | 0.02 |
ENSMUST00000197642.2
ENSMUST00000026895.14 ENSMUST00000169922.9 |
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr16_-_92622659 | 0.02 |
ENSMUST00000186296.2
|
Runx1
|
runt related transcription factor 1 |
| chr2_+_174126103 | 0.02 |
ENSMUST00000109095.8
ENSMUST00000180362.8 ENSMUST00000109096.9 |
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr11_-_77616103 | 0.02 |
ENSMUST00000078623.5
|
Cryba1
|
crystallin, beta A1 |
| chr7_-_101859033 | 0.02 |
ENSMUST00000211005.2
|
Nup98
|
nucleoporin 98 |
| chr7_+_18810167 | 0.02 |
ENSMUST00000108479.2
|
Dmwd
|
dystrophia myotonica-containing WD repeat motif |
| chr9_+_102593871 | 0.02 |
ENSMUST00000145913.2
ENSMUST00000153965.8 |
Amotl2
|
angiomotin-like 2 |
| chr7_-_110462446 | 0.02 |
ENSMUST00000033050.5
|
Lyve1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr2_-_25982160 | 0.02 |
ENSMUST00000114159.9
|
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing |
| chr4_+_11558905 | 0.02 |
ENSMUST00000095145.12
ENSMUST00000108306.9 ENSMUST00000070755.13 |
Rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr9_+_119723931 | 0.02 |
ENSMUST00000036561.8
ENSMUST00000217472.2 ENSMUST00000215307.2 |
Wdr48
|
WD repeat domain 48 |
| chr6_-_113354468 | 0.02 |
ENSMUST00000099118.8
|
Tada3
|
transcriptional adaptor 3 |
| chr9_-_20864096 | 0.02 |
ENSMUST00000004202.17
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1 |
| chr6_-_113354337 | 0.02 |
ENSMUST00000043333.9
|
Tada3
|
transcriptional adaptor 3 |
| chr17_-_23959334 | 0.02 |
ENSMUST00000024702.5
|
Paqr4
|
progestin and adipoQ receptor family member IV |
| chr6_+_86055018 | 0.02 |
ENSMUST00000205034.3
ENSMUST00000203724.3 |
Add2
|
adducin 2 (beta) |
| chr14_+_27057980 | 0.02 |
ENSMUST00000224981.2
|
Arhgef3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr13_+_52750883 | 0.02 |
ENSMUST00000055087.7
|
Syk
|
spleen tyrosine kinase |
| chr5_-_99876898 | 0.02 |
ENSMUST00000146396.8
ENSMUST00000161148.2 ENSMUST00000161516.2 |
Rasgef1b
|
RasGEF domain family, member 1B |
| chr17_-_48235385 | 0.02 |
ENSMUST00000113262.2
|
Foxp4
|
forkhead box P4 |
| chr10_-_81186025 | 0.02 |
ENSMUST00000122993.8
|
Hmg20b
|
high mobility group 20B |
| chr4_-_130068484 | 0.02 |
ENSMUST00000132545.3
ENSMUST00000175992.8 ENSMUST00000105999.9 |
Tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp784
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0002545 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.1 | 0.2 | GO:1904431 | response to intra-S DNA damage checkpoint signaling(GO:0072429) positive regulation of t-circle formation(GO:1904431) |
| 0.1 | 0.2 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.1 | 0.3 | GO:0002358 | pre-B cell allelic exclusion(GO:0002331) B cell homeostatic proliferation(GO:0002358) |
| 0.1 | 0.5 | GO:0098735 | cellular response to caffeine(GO:0071313) positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:2000097 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:1901204 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.0 | 0.2 | GO:0050760 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:1902365 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.1 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 0.2 | GO:0090274 | reduction of food intake in response to dietary excess(GO:0002023) positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 | 0.2 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.2 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.0 | 0.5 | GO:0045060 | negative thymic T cell selection(GO:0045060) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.2 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.5 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.1 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.0 | 0.2 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0060464 | lung lobe formation(GO:0060464) diaphragm morphogenesis(GO:0060540) |
| 0.0 | 0.3 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.0 | 0.2 | GO:1901250 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.0 | 0.0 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.2 | GO:1904668 | mitotic spindle midzone assembly(GO:0051256) positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.2 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.2 | GO:0048312 | inositol phosphate dephosphorylation(GO:0046855) intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.2 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.1 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.2 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.0 | GO:1904602 | serotonin-activated cation-selective channel complex(GO:1904602) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.3 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.1 | 0.5 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.2 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.1 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |