Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
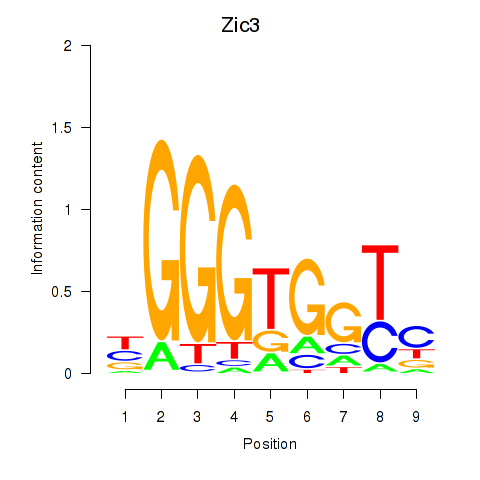
Results for Zic3
Z-value: 0.31
Transcription factors associated with Zic3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zic3
|
ENSMUSG00000067860.12 | zinc finger protein of the cerebellum 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zic3 | mm39_v1_chrX_+_57076359_57076378 | 0.02 | 9.8e-01 | Click! |
Activity profile of Zic3 motif
Sorted Z-values of Zic3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_115790768 | 0.25 |
ENSMUST00000152171.8
|
Smim5
|
small integral membrane protein 5 |
| chr6_-_52211882 | 0.15 |
ENSMUST00000125581.2
|
Hoxa10
|
homeobox A10 |
| chr15_+_78912642 | 0.15 |
ENSMUST00000180086.3
|
H1f0
|
H1.0 linker histone |
| chr6_-_52168675 | 0.13 |
ENSMUST00000101395.3
|
Hoxa4
|
homeobox A4 |
| chr9_+_21746785 | 0.13 |
ENSMUST00000058777.8
|
Angptl8
|
angiopoietin-like 8 |
| chr16_-_4376471 | 0.11 |
ENSMUST00000230875.2
|
Tfap4
|
transcription factor AP4 |
| chr10_-_128245501 | 0.11 |
ENSMUST00000172348.8
ENSMUST00000166608.8 ENSMUST00000164199.8 ENSMUST00000171370.2 ENSMUST00000026439.14 |
Nabp2
|
nucleic acid binding protein 2 |
| chr10_-_49664839 | 0.10 |
ENSMUST00000220263.2
ENSMUST00000218823.2 |
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2) |
| chr12_-_31549538 | 0.10 |
ENSMUST00000064240.14
ENSMUST00000185739.8 ENSMUST00000188326.3 ENSMUST00000101499.10 ENSMUST00000085487.12 |
Cbll1
|
Casitas B-lineage lymphoma-like 1 |
| chr11_-_70545450 | 0.09 |
ENSMUST00000018437.3
|
Pfn1
|
profilin 1 |
| chr11_-_69304501 | 0.09 |
ENSMUST00000094077.5
|
Kdm6b
|
KDM1 lysine (K)-specific demethylase 6B |
| chr2_+_28082943 | 0.09 |
ENSMUST00000113920.8
|
Olfm1
|
olfactomedin 1 |
| chr9_+_106247943 | 0.09 |
ENSMUST00000173748.2
|
Dusp7
|
dual specificity phosphatase 7 |
| chr10_-_95159933 | 0.09 |
ENSMUST00000053594.7
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr15_-_101268036 | 0.09 |
ENSMUST00000077196.6
|
Krt80
|
keratin 80 |
| chr10_-_95159410 | 0.08 |
ENSMUST00000217809.2
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr5_-_107873883 | 0.08 |
ENSMUST00000159263.3
|
Gfi1
|
growth factor independent 1 transcription repressor |
| chr5_-_66309244 | 0.08 |
ENSMUST00000167950.8
|
Rbm47
|
RNA binding motif protein 47 |
| chr11_+_115790951 | 0.08 |
ENSMUST00000142089.2
ENSMUST00000131566.2 |
Smim5
|
small integral membrane protein 5 |
| chr5_-_66308421 | 0.08 |
ENSMUST00000200775.4
ENSMUST00000094756.11 |
Rbm47
|
RNA binding motif protein 47 |
| chr16_-_89757693 | 0.07 |
ENSMUST00000002588.11
|
Tiam1
|
T cell lymphoma invasion and metastasis 1 |
| chr2_-_170269748 | 0.07 |
ENSMUST00000013667.3
ENSMUST00000109152.9 ENSMUST00000068137.11 |
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr13_+_44882998 | 0.07 |
ENSMUST00000174068.8
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr7_+_127078371 | 0.07 |
ENSMUST00000205432.3
|
Fbrs
|
fibrosin |
| chr11_+_119158713 | 0.07 |
ENSMUST00000106259.9
|
Gaa
|
glucosidase, alpha, acid |
| chr11_+_72889889 | 0.06 |
ENSMUST00000021141.14
|
P2rx1
|
purinergic receptor P2X, ligand-gated ion channel, 1 |
| chr7_+_127399789 | 0.06 |
ENSMUST00000125188.8
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr2_+_90865958 | 0.06 |
ENSMUST00000111445.10
ENSMUST00000111446.10 ENSMUST00000050323.6 |
Rapsn
|
receptor-associated protein of the synapse |
| chr4_-_62126662 | 0.06 |
ENSMUST00000222050.2
ENSMUST00000068822.4 |
Zfp37
|
zinc finger protein 37 |
| chr7_+_18618605 | 0.06 |
ENSMUST00000032573.8
|
Pglyrp1
|
peptidoglycan recognition protein 1 |
| chr6_-_124733441 | 0.06 |
ENSMUST00000088357.12
|
Atn1
|
atrophin 1 |
| chr7_-_48530777 | 0.06 |
ENSMUST00000058745.15
|
E2f8
|
E2F transcription factor 8 |
| chr5_-_66308666 | 0.06 |
ENSMUST00000201561.4
|
Rbm47
|
RNA binding motif protein 47 |
| chr10_-_30679289 | 0.06 |
ENSMUST00000215725.2
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr7_+_45522196 | 0.06 |
ENSMUST00000002855.14
ENSMUST00000211716.2 |
Kdelr1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr5_-_113957318 | 0.06 |
ENSMUST00000201194.4
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr9_-_88320937 | 0.06 |
ENSMUST00000173011.9
|
Snx14
|
sorting nexin 14 |
| chr5_-_145077048 | 0.06 |
ENSMUST00000031627.9
|
Pdap1
|
PDGFA associated protein 1 |
| chr6_-_146403638 | 0.06 |
ENSMUST00000079573.13
|
Itpr2
|
inositol 1,4,5-triphosphate receptor 2 |
| chr11_+_69214895 | 0.06 |
ENSMUST00000060956.13
|
Trappc1
|
trafficking protein particle complex 1 |
| chr8_+_120163857 | 0.06 |
ENSMUST00000152420.8
ENSMUST00000212112.2 ENSMUST00000098365.4 |
Osgin1
|
oxidative stress induced growth inhibitor 1 |
| chr14_-_64654397 | 0.06 |
ENSMUST00000210428.2
|
Msra
|
methionine sulfoxide reductase A |
| chr6_-_39702127 | 0.05 |
ENSMUST00000101497.4
|
Braf
|
Braf transforming gene |
| chr18_+_7869705 | 0.05 |
ENSMUST00000166062.8
ENSMUST00000169010.8 |
Wac
|
WW domain containing adaptor with coiled-coil |
| chr6_+_29531247 | 0.05 |
ENSMUST00000164922.3
|
Irf5
|
interferon regulatory factor 5 |
| chr1_-_182109773 | 0.05 |
ENSMUST00000133052.2
|
Degs1
|
delta(4)-desaturase, sphingolipid 1 |
| chr12_+_112727089 | 0.05 |
ENSMUST00000063888.5
|
Pld4
|
phospholipase D family, member 4 |
| chr4_+_123798690 | 0.05 |
ENSMUST00000106202.4
|
Mycbp
|
MYC binding protein |
| chr11_+_77985508 | 0.05 |
ENSMUST00000100782.4
|
Fam222b
|
family with sequence similarity 222, member B |
| chr1_-_171910324 | 0.05 |
ENSMUST00000003550.11
|
Ncstn
|
nicastrin |
| chr3_-_144425819 | 0.05 |
ENSMUST00000199531.5
ENSMUST00000199854.5 |
Sh3glb1
|
SH3-domain GRB2-like B1 (endophilin) |
| chr1_-_105284383 | 0.05 |
ENSMUST00000058688.7
|
Rnf152
|
ring finger protein 152 |
| chr2_+_121287444 | 0.05 |
ENSMUST00000126764.2
|
Hypk
|
huntingtin interacting protein K |
| chrX_+_162691978 | 0.05 |
ENSMUST00000069041.15
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr1_+_74275683 | 0.05 |
ENSMUST00000113819.2
|
Arpc2
|
actin related protein 2/3 complex, subunit 2 |
| chr9_-_88320991 | 0.05 |
ENSMUST00000239462.2
ENSMUST00000165315.9 ENSMUST00000173039.9 |
Snx14
|
sorting nexin 14 |
| chr15_-_98851423 | 0.05 |
ENSMUST00000134214.3
|
Gm49450
|
predicted gene, 49450 |
| chr18_-_78185334 | 0.05 |
ENSMUST00000160639.2
|
Slc14a1
|
solute carrier family 14 (urea transporter), member 1 |
| chr17_-_71617945 | 0.04 |
ENSMUST00000232777.2
ENSMUST00000024849.11 |
Emilin2
|
elastin microfibril interfacer 2 |
| chr17_-_71617968 | 0.04 |
ENSMUST00000233057.2
|
Emilin2
|
elastin microfibril interfacer 2 |
| chr3_+_63203235 | 0.04 |
ENSMUST00000194134.6
|
Mme
|
membrane metallo endopeptidase |
| chr17_+_35354148 | 0.04 |
ENSMUST00000166426.9
ENSMUST00000025250.14 |
Bag6
|
BCL2-associated athanogene 6 |
| chr1_-_105284407 | 0.04 |
ENSMUST00000172299.2
|
Rnf152
|
ring finger protein 152 |
| chr9_-_110474398 | 0.04 |
ENSMUST00000149089.2
|
Nbeal2
|
neurobeachin-like 2 |
| chr2_+_143757193 | 0.04 |
ENSMUST00000103172.4
|
Dstn
|
destrin |
| chr14_-_104081119 | 0.04 |
ENSMUST00000227824.2
ENSMUST00000172237.2 |
Ednrb
|
endothelin receptor type B |
| chr8_-_71213535 | 0.04 |
ENSMUST00000038626.10
|
Mpv17l2
|
MPV17 mitochondrial membrane protein-like 2 |
| chr1_-_177624017 | 0.04 |
ENSMUST00000016105.9
|
Adss
|
adenylosuccinate synthetase, non muscle |
| chr7_-_126398343 | 0.04 |
ENSMUST00000032934.12
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr4_-_53159885 | 0.04 |
ENSMUST00000030010.4
|
Abca1
|
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chr17_+_35354655 | 0.04 |
ENSMUST00000174478.8
ENSMUST00000174281.9 ENSMUST00000173550.8 |
Bag6
|
BCL2-associated athanogene 6 |
| chr10_-_127358231 | 0.04 |
ENSMUST00000219239.2
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr17_+_34457868 | 0.04 |
ENSMUST00000095342.11
ENSMUST00000167280.8 ENSMUST00000236838.2 |
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr16_-_85347305 | 0.04 |
ENSMUST00000175700.8
ENSMUST00000114174.3 |
Cyyr1
|
cysteine and tyrosine-rich protein 1 |
| chr6_+_146479357 | 0.04 |
ENSMUST00000067404.13
ENSMUST00000111663.9 ENSMUST00000058245.5 |
Fgfr1op2
|
FGFR1 oncogene partner 2 |
| chr6_-_52195663 | 0.04 |
ENSMUST00000134367.4
|
Hoxa7
|
homeobox A7 |
| chr11_+_77985486 | 0.04 |
ENSMUST00000073705.12
|
Fam222b
|
family with sequence similarity 222, member B |
| chr13_+_74269554 | 0.04 |
ENSMUST00000036208.7
ENSMUST00000225423.2 ENSMUST00000221703.2 |
Slc9a3
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 |
| chr9_+_56979307 | 0.04 |
ENSMUST00000169879.8
|
Sin3a
|
transcriptional regulator, SIN3A (yeast) |
| chr17_+_35354430 | 0.04 |
ENSMUST00000173535.8
ENSMUST00000173952.8 |
Bag6
|
BCL2-associated athanogene 6 |
| chr6_-_146479323 | 0.03 |
ENSMUST00000032427.15
|
Ints13
|
integrator complex subunit 13 |
| chrX_+_67722390 | 0.03 |
ENSMUST00000114654.8
ENSMUST00000114655.8 ENSMUST00000114657.9 ENSMUST00000114653.2 |
Fmr1
|
FMRP translational regulator 1 |
| chr11_-_101169753 | 0.03 |
ENSMUST00000168089.2
ENSMUST00000017332.4 |
Coa3
|
cytochrome C oxidase assembly factor 3 |
| chr3_+_132389867 | 0.03 |
ENSMUST00000169172.5
|
Tbck
|
TBC1 domain containing kinase |
| chr12_+_3622379 | 0.03 |
ENSMUST00000173199.8
ENSMUST00000164578.9 ENSMUST00000174479.8 ENSMUST00000173240.8 ENSMUST00000174663.8 ENSMUST00000173736.8 |
Dtnb
|
dystrobrevin, beta |
| chr2_+_156563284 | 0.03 |
ENSMUST00000099145.6
|
Dlgap4
|
DLG associated protein 4 |
| chr11_+_106265660 | 0.03 |
ENSMUST00000188561.7
ENSMUST00000190795.7 ENSMUST00000185986.7 ENSMUST00000190268.2 |
Prr29
|
proline rich 29 |
| chrX_-_107877909 | 0.03 |
ENSMUST00000101283.4
ENSMUST00000150434.8 |
Brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr1_-_123973223 | 0.03 |
ENSMUST00000112606.8
|
Dpp10
|
dipeptidylpeptidase 10 |
| chr17_+_35354172 | 0.03 |
ENSMUST00000172571.8
ENSMUST00000173491.8 |
Bag6
|
BCL2-associated athanogene 6 |
| chr7_-_80453033 | 0.03 |
ENSMUST00000167377.3
|
Iqgap1
|
IQ motif containing GTPase activating protein 1 |
| chr7_+_127400016 | 0.03 |
ENSMUST00000106271.2
ENSMUST00000138432.2 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr7_-_126303947 | 0.03 |
ENSMUST00000032949.14
|
Coro1a
|
coronin, actin binding protein 1A |
| chr8_+_127790772 | 0.03 |
ENSMUST00000079777.12
ENSMUST00000160272.8 ENSMUST00000162907.8 ENSMUST00000162536.8 ENSMUST00000026921.13 ENSMUST00000162665.8 ENSMUST00000162602.8 ENSMUST00000160581.8 ENSMUST00000161355.8 ENSMUST00000162531.8 ENSMUST00000160766.8 ENSMUST00000159537.8 |
Pard3
|
par-3 family cell polarity regulator |
| chr3_+_53371086 | 0.03 |
ENSMUST00000058577.5
|
Proser1
|
proline and serine rich 1 |
| chr4_+_123798625 | 0.03 |
ENSMUST00000030400.14
|
Mycbp
|
MYC binding protein |
| chr3_-_108322868 | 0.03 |
ENSMUST00000090558.10
|
Celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr12_+_81678143 | 0.03 |
ENSMUST00000036116.6
|
Ttc9
|
tetratricopeptide repeat domain 9 |
| chr9_-_56325344 | 0.03 |
ENSMUST00000061552.15
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr17_-_24428351 | 0.03 |
ENSMUST00000024931.6
|
Ntn3
|
netrin 3 |
| chr9_-_88320962 | 0.03 |
ENSMUST00000174806.9
|
Snx14
|
sorting nexin 14 |
| chr7_+_127399776 | 0.03 |
ENSMUST00000046863.12
ENSMUST00000206674.2 ENSMUST00000106272.8 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr1_-_155848917 | 0.03 |
ENSMUST00000138762.8
|
Cep350
|
centrosomal protein 350 |
| chrX_+_10583629 | 0.03 |
ENSMUST00000115524.8
ENSMUST00000008179.7 ENSMUST00000156321.2 |
Mid1ip1
|
Mid1 interacting protein 1 (gastrulation specific G12-like (zebrafish)) |
| chr6_+_90596123 | 0.03 |
ENSMUST00000032177.10
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr1_+_82817794 | 0.03 |
ENSMUST00000186043.2
|
Agfg1
|
ArfGAP with FG repeats 1 |
| chr10_-_127358300 | 0.03 |
ENSMUST00000026470.6
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr12_+_76593799 | 0.03 |
ENSMUST00000218380.2
ENSMUST00000219751.2 |
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr7_+_78563964 | 0.03 |
ENSMUST00000120331.4
|
Isg20
|
interferon-stimulated protein |
| chr5_-_136595205 | 0.03 |
ENSMUST00000176778.8
|
Cux1
|
cut-like homeobox 1 |
| chr16_+_17327076 | 0.03 |
ENSMUST00000232242.2
|
Lztr1
|
leucine-zipper-like transcriptional regulator, 1 |
| chr1_+_82817388 | 0.03 |
ENSMUST00000190052.7
ENSMUST00000063380.11 ENSMUST00000187899.7 ENSMUST00000186302.7 ENSMUST00000190046.7 |
Agfg1
|
ArfGAP with FG repeats 1 |
| chr9_-_108141105 | 0.03 |
ENSMUST00000166905.8
ENSMUST00000191899.6 |
Dag1
|
dystroglycan 1 |
| chr5_-_113957362 | 0.03 |
ENSMUST00000202555.2
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr4_+_140688514 | 0.03 |
ENSMUST00000010007.9
|
Sdhb
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr16_+_32151056 | 0.03 |
ENSMUST00000115151.5
ENSMUST00000232137.2 |
Ubxn7
|
UBX domain protein 7 |
| chr2_+_29759495 | 0.03 |
ENSMUST00000047521.7
ENSMUST00000134152.2 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chr17_+_47906985 | 0.03 |
ENSMUST00000182539.8
|
Ccnd3
|
cyclin D3 |
| chr1_+_74401267 | 0.03 |
ENSMUST00000097697.8
|
Catip
|
ciliogenesis associated TTC17 interacting protein |
| chr12_-_108801763 | 0.03 |
ENSMUST00000021693.4
|
Slc25a29
|
solute carrier family 25 (mitochondrial carrier, palmitoylcarnitine transporter), member 29 |
| chr17_+_56312672 | 0.03 |
ENSMUST00000133998.8
|
Mpnd
|
MPN domain containing |
| chr11_-_78642480 | 0.03 |
ENSMUST00000059468.6
|
Ccnq
|
cyclin Q |
| chr19_-_4889284 | 0.03 |
ENSMUST00000236451.2
ENSMUST00000236178.2 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr11_+_98727611 | 0.03 |
ENSMUST00000107479.3
|
Rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr15_-_66158445 | 0.03 |
ENSMUST00000070256.9
|
Kcnq3
|
potassium voltage-gated channel, subfamily Q, member 3 |
| chr10_-_43416941 | 0.03 |
ENSMUST00000147196.3
|
Mtres1
|
mitochondrial transcription rescue factor 1 |
| chr11_+_69806866 | 0.03 |
ENSMUST00000134581.2
|
Gps2
|
G protein pathway suppressor 2 |
| chr18_-_67857594 | 0.02 |
ENSMUST00000120934.8
ENSMUST00000025420.14 ENSMUST00000122412.2 |
Ptpn2
|
protein tyrosine phosphatase, non-receptor type 2 |
| chr9_-_44632680 | 0.02 |
ENSMUST00000148929.2
ENSMUST00000123406.8 |
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr11_-_72380730 | 0.02 |
ENSMUST00000045303.10
|
Spns2
|
spinster homolog 2 |
| chr6_-_39702381 | 0.02 |
ENSMUST00000002487.15
|
Braf
|
Braf transforming gene |
| chr4_-_70453140 | 0.02 |
ENSMUST00000107359.9
|
Megf9
|
multiple EGF-like-domains 9 |
| chr10_+_106306122 | 0.02 |
ENSMUST00000029404.17
ENSMUST00000217854.2 |
Ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr18_-_9282754 | 0.02 |
ENSMUST00000041007.4
|
Gjd4
|
gap junction protein, delta 4 |
| chr9_+_90045219 | 0.02 |
ENSMUST00000147250.8
ENSMUST00000113060.3 |
Adamts7
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 7 |
| chr9_+_106331041 | 0.02 |
ENSMUST00000024260.14
ENSMUST00000216379.2 ENSMUST00000215656.2 ENSMUST00000214252.2 |
Pcbp4
|
poly(rC) binding protein 4 |
| chr1_+_192835414 | 0.02 |
ENSMUST00000076521.7
|
Irf6
|
interferon regulatory factor 6 |
| chr4_-_133066549 | 0.02 |
ENSMUST00000105906.2
|
Wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr17_-_85397627 | 0.02 |
ENSMUST00000072406.5
ENSMUST00000171795.9 ENSMUST00000234676.2 |
Prepl
|
prolyl endopeptidase-like |
| chr6_+_116241146 | 0.02 |
ENSMUST00000112900.9
ENSMUST00000036503.14 ENSMUST00000223495.2 |
Zfand4
|
zinc finger, AN1-type domain 4 |
| chr7_+_127399848 | 0.02 |
ENSMUST00000139068.8
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr3_+_89136353 | 0.02 |
ENSMUST00000041142.4
|
Muc1
|
mucin 1, transmembrane |
| chr6_-_13677928 | 0.02 |
ENSMUST00000203078.2
ENSMUST00000045235.8 |
Bmt2
|
base methyltransferase of 25S rRNA 2 |
| chr7_-_126992776 | 0.02 |
ENSMUST00000165495.2
ENSMUST00000106303.3 ENSMUST00000074249.7 |
E430018J23Rik
|
RIKEN cDNA E430018J23 gene |
| chr14_+_54704942 | 0.02 |
ENSMUST00000228407.2
|
Lrp10
|
low-density lipoprotein receptor-related protein 10 |
| chr6_-_99643723 | 0.02 |
ENSMUST00000032151.3
|
Eif4e3
|
eukaryotic translation initiation factor 4E member 3 |
| chr11_-_70590923 | 0.02 |
ENSMUST00000108543.4
ENSMUST00000108542.8 ENSMUST00000108541.9 ENSMUST00000126114.9 ENSMUST00000073625.8 |
Inca1
|
inhibitor of CDK, cyclin A1 interacting protein 1 |
| chrX_+_7628891 | 0.02 |
ENSMUST00000077680.10
ENSMUST00000079542.13 ENSMUST00000115679.8 ENSMUST00000137467.8 |
Tfe3
|
transcription factor E3 |
| chr1_-_119349969 | 0.02 |
ENSMUST00000038765.6
|
Inhbb
|
inhibin beta-B |
| chr15_+_78294154 | 0.02 |
ENSMUST00000229739.2
|
Mpst
|
mercaptopyruvate sulfurtransferase |
| chr11_+_77384234 | 0.02 |
ENSMUST00000037285.10
ENSMUST00000100812.4 |
Git1
|
GIT ArfGAP 1 |
| chr12_+_81678002 | 0.02 |
ENSMUST00000218362.2
|
Ttc9
|
tetratricopeptide repeat domain 9 |
| chr16_+_48662894 | 0.02 |
ENSMUST00000238847.2
ENSMUST00000023329.7 |
Retnla
|
resistin like alpha |
| chr15_+_74435587 | 0.02 |
ENSMUST00000185682.7
ENSMUST00000170845.8 ENSMUST00000187599.2 |
Adgrb1
|
adhesion G protein-coupled receptor B1 |
| chr1_+_129201081 | 0.02 |
ENSMUST00000073527.13
ENSMUST00000040311.14 |
Thsd7b
|
thrombospondin, type I, domain containing 7B |
| chr5_-_123859070 | 0.02 |
ENSMUST00000031376.12
|
Zcchc8
|
zinc finger, CCHC domain containing 8 |
| chr15_-_99717956 | 0.02 |
ENSMUST00000109024.9
|
Lima1
|
LIM domain and actin binding 1 |
| chr7_-_105230395 | 0.02 |
ENSMUST00000188726.2
ENSMUST00000188440.7 |
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr4_+_20042045 | 0.02 |
ENSMUST00000098242.4
|
Ggh
|
gamma-glutamyl hydrolase |
| chr9_+_65967477 | 0.02 |
ENSMUST00000034947.7
|
Ppib
|
peptidylprolyl isomerase B |
| chr8_+_106587212 | 0.02 |
ENSMUST00000008594.9
|
Nutf2
|
nuclear transport factor 2 |
| chr2_-_148250024 | 0.02 |
ENSMUST00000099270.5
|
Thbd
|
thrombomodulin |
| chr14_+_34097422 | 0.02 |
ENSMUST00000111908.3
|
Mmrn2
|
multimerin 2 |
| chr2_+_70392351 | 0.02 |
ENSMUST00000094934.11
|
Gad1
|
glutamate decarboxylase 1 |
| chrX_-_162332673 | 0.02 |
ENSMUST00000033730.3
|
Grpr
|
gastrin releasing peptide receptor |
| chr5_-_24963006 | 0.02 |
ENSMUST00000047119.5
|
Crygn
|
crystallin, gamma N |
| chr2_+_131075965 | 0.02 |
ENSMUST00000041362.12
ENSMUST00000110199.3 |
Mavs
|
mitochondrial antiviral signaling protein |
| chr10_+_71183657 | 0.02 |
ENSMUST00000118381.8
ENSMUST00000121446.2 |
Ipmk
|
inositol polyphosphate multikinase |
| chr11_+_102556397 | 0.02 |
ENSMUST00000100378.4
|
Meioc
|
meiosis specific with coiled-coil domain |
| chr3_+_90161470 | 0.02 |
ENSMUST00000029545.15
|
Crtc2
|
CREB regulated transcription coactivator 2 |
| chr11_-_121279062 | 0.02 |
ENSMUST00000106107.3
|
Rab40b
|
Rab40B, member RAS oncogene family |
| chr9_-_54956032 | 0.02 |
ENSMUST00000034854.8
|
Chrnb4
|
cholinergic receptor, nicotinic, beta polypeptide 4 |
| chr17_+_21165573 | 0.02 |
ENSMUST00000007708.14
|
Ppp2r1a
|
protein phosphatase 2, regulatory subunit A, alpha |
| chr11_+_100751272 | 0.02 |
ENSMUST00000107357.4
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr6_+_48372520 | 0.02 |
ENSMUST00000114571.8
ENSMUST00000031815.12 ENSMUST00000203371.3 |
Krba1
|
KRAB-A domain containing 1 |
| chr2_-_148285450 | 0.02 |
ENSMUST00000099269.4
|
Cd93
|
CD93 antigen |
| chr7_-_4999099 | 0.02 |
ENSMUST00000108572.2
|
Zfp579
|
zinc finger protein 579 |
| chr1_+_135075377 | 0.02 |
ENSMUST00000125774.2
|
Arl8a
|
ADP-ribosylation factor-like 8A |
| chr14_-_73563212 | 0.02 |
ENSMUST00000022701.7
|
Rb1
|
RB transcriptional corepressor 1 |
| chr9_-_64644905 | 0.02 |
ENSMUST00000169058.8
|
Rab11a
|
RAB11A, member RAS oncogene family |
| chr17_+_34134873 | 0.02 |
ENSMUST00000172619.8
ENSMUST00000174463.2 |
Tapbp
Zbtb22
|
TAP binding protein zinc finger and BTB domain containing 22 |
| chr6_-_51446752 | 0.02 |
ENSMUST00000204188.3
ENSMUST00000203220.3 ENSMUST00000114459.8 ENSMUST00000090002.10 |
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr6_+_73225616 | 0.02 |
ENSMUST00000203632.2
|
Suclg1
|
succinate-CoA ligase, GDP-forming, alpha subunit |
| chr10_-_62859661 | 0.02 |
ENSMUST00000020263.14
ENSMUST00000118898.8 |
Hnrnph3
|
heterogeneous nuclear ribonucleoprotein H3 |
| chr12_-_21423551 | 0.02 |
ENSMUST00000101551.10
|
Adam17
|
a disintegrin and metallopeptidase domain 17 |
| chr7_-_111379170 | 0.02 |
ENSMUST00000049430.15
ENSMUST00000106663.2 |
Galnt18
|
polypeptide N-acetylgalactosaminyltransferase 18 |
| chr11_-_69696428 | 0.01 |
ENSMUST00000051025.5
|
Tmem102
|
transmembrane protein 102 |
| chr12_-_21423607 | 0.01 |
ENSMUST00000064536.13
|
Adam17
|
a disintegrin and metallopeptidase domain 17 |
| chr11_+_6241606 | 0.01 |
ENSMUST00000003461.15
|
Ogdh
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr15_+_102178975 | 0.01 |
ENSMUST00000181801.2
|
Gm9918
|
predicted gene 9918 |
| chr19_+_5928649 | 0.01 |
ENSMUST00000136833.8
ENSMUST00000141362.2 |
Slc25a45
|
solute carrier family 25, member 45 |
| chr11_-_102837514 | 0.01 |
ENSMUST00000057849.6
|
C1ql1
|
complement component 1, q subcomponent-like 1 |
| chr2_-_30871875 | 0.01 |
ENSMUST00000028205.10
|
BC005624
|
cDNA sequence BC005624 |
| chr8_+_105267431 | 0.01 |
ENSMUST00000056051.11
|
Car7
|
carbonic anhydrase 7 |
| chr1_-_13444249 | 0.01 |
ENSMUST00000068304.13
ENSMUST00000006037.13 |
Ncoa2
|
nuclear receptor coactivator 2 |
| chr2_+_53082079 | 0.01 |
ENSMUST00000028336.7
|
Arl6ip6
|
ADP-ribosylation factor-like 6 interacting protein 6 |
| chr2_-_155953783 | 0.01 |
ENSMUST00000154889.8
ENSMUST00000079312.10 ENSMUST00000153634.8 ENSMUST00000133921.8 ENSMUST00000184265.8 ENSMUST00000109607.10 ENSMUST00000109608.9 ENSMUST00000131377.8 ENSMUST00000183518.2 |
Cpne1
Rbm12
Gm28036
|
copine I RNA binding motif protein 12 predicted gene, 28036 |
| chr7_+_98917542 | 0.01 |
ENSMUST00000107100.3
ENSMUST00000208605.2 |
Map6
|
microtubule-associated protein 6 |
| chr3_+_138058139 | 0.01 |
ENSMUST00000090166.5
|
Adh6b
|
alcohol dehydrogenase 6B (class V) |
| chr14_+_54713557 | 0.01 |
ENSMUST00000164766.8
|
Rem2
|
rad and gem related GTP binding protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zic3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.0 | 0.1 | GO:1904266 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.0 | 0.1 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.1 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.0 | 0.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.1 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.1 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.0 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.0 | 0.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.0 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0002554 | serotonin secretion by platelet(GO:0002554) |
| 0.0 | 0.0 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.0 | 0.1 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.0 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.0 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.0 | GO:0034188 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.1 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |