Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
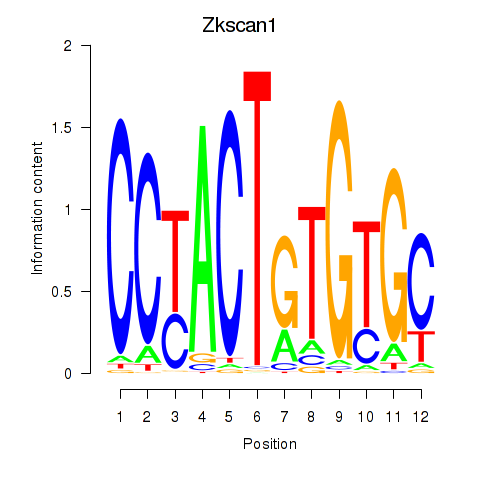
Results for Zkscan1
Z-value: 0.58
Transcription factors associated with Zkscan1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zkscan1
|
ENSMUSG00000029729.13 | zinc finger with KRAB and SCAN domains 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zkscan1 | mm39_v1_chr5_+_138083345_138083410 | -0.79 | 1.1e-01 | Click! |
Activity profile of Zkscan1 motif
Sorted Z-values of Zkscan1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_74920518 | 0.36 |
ENSMUST00000185372.2
ENSMUST00000187347.7 ENSMUST00000188845.7 ENSMUST00000185200.7 ENSMUST00000179762.8 ENSMUST00000191216.7 ENSMUST00000065408.16 |
Ly6c1
|
lymphocyte antigen 6 complex, locus C1 |
| chr15_-_74983786 | 0.28 |
ENSMUST00000191451.2
ENSMUST00000100542.10 |
Ly6c2
|
lymphocyte antigen 6 complex, locus C2 |
| chr2_+_31135813 | 0.27 |
ENSMUST00000000199.8
|
Ncs1
|
neuronal calcium sensor 1 |
| chr19_-_61216834 | 0.24 |
ENSMUST00000076046.7
|
Csf2ra
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr11_-_78313043 | 0.24 |
ENSMUST00000001122.6
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr15_-_74855264 | 0.23 |
ENSMUST00000023250.11
ENSMUST00000166694.2 |
Ly6i
|
lymphocyte antigen 6 complex, locus I |
| chr19_+_53298906 | 0.20 |
ENSMUST00000003870.15
|
Mxi1
|
MAX interactor 1, dimerization protein |
| chr4_+_45890303 | 0.20 |
ENSMUST00000178561.8
|
Ccdc180
|
coiled-coil domain containing 180 |
| chr15_-_74869684 | 0.19 |
ENSMUST00000190188.2
ENSMUST00000189068.7 ENSMUST00000186526.7 ENSMUST00000187171.2 ENSMUST00000187994.7 |
Ly6a
|
lymphocyte antigen 6 complex, locus A |
| chr9_+_62249177 | 0.19 |
ENSMUST00000128636.8
|
Anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr16_-_17019321 | 0.18 |
ENSMUST00000232668.2
ENSMUST00000231643.2 ENSMUST00000232035.2 |
Ube2l3
|
ubiquitin-conjugating enzyme E2L 3 |
| chr9_+_62249341 | 0.18 |
ENSMUST00000135395.8
|
Anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr1_+_88022776 | 0.18 |
ENSMUST00000150634.8
ENSMUST00000058237.14 |
Ugt1a7c
|
UDP glucuronosyltransferase 1 family, polypeptide A7C |
| chr16_-_17019352 | 0.18 |
ENSMUST00000090192.12
|
Ube2l3
|
ubiquitin-conjugating enzyme E2L 3 |
| chr15_+_75027089 | 0.18 |
ENSMUST00000190262.2
|
Ly6g
|
lymphocyte antigen 6 complex, locus G |
| chr12_+_112586501 | 0.16 |
ENSMUST00000180015.9
|
Adssl1
|
adenylosuccinate synthetase like 1 |
| chr10_+_79716876 | 0.16 |
ENSMUST00000166201.2
|
Prtn3
|
proteinase 3 |
| chr2_+_112092271 | 0.16 |
ENSMUST00000028553.4
|
Nop10
|
NOP10 ribonucleoprotein |
| chr8_+_95712151 | 0.15 |
ENSMUST00000212799.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr9_+_62248611 | 0.15 |
ENSMUST00000085519.13
|
Anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr15_-_74869483 | 0.15 |
ENSMUST00000023248.13
|
Ly6a
|
lymphocyte antigen 6 complex, locus A |
| chr12_+_112586465 | 0.15 |
ENSMUST00000021726.8
|
Adssl1
|
adenylosuccinate synthetase like 1 |
| chr4_-_63414188 | 0.13 |
ENSMUST00000063650.10
ENSMUST00000102867.8 ENSMUST00000107393.8 ENSMUST00000084510.8 ENSMUST00000095038.8 ENSMUST00000119294.8 ENSMUST00000095037.2 ENSMUST00000063672.10 |
Whrn
|
whirlin |
| chr11_-_102588536 | 0.13 |
ENSMUST00000164506.3
ENSMUST00000092569.13 |
Ccdc43
|
coiled-coil domain containing 43 |
| chr1_+_171668173 | 0.11 |
ENSMUST00000136479.8
|
Cd84
|
CD84 antigen |
| chr1_+_90531183 | 0.11 |
ENSMUST00000186750.2
|
Cops8
|
COP9 signalosome subunit 8 |
| chr10_+_126836578 | 0.10 |
ENSMUST00000026500.12
ENSMUST00000142698.8 |
Avil
|
advillin |
| chr2_+_172235820 | 0.10 |
ENSMUST00000109136.3
ENSMUST00000228775.2 |
Cass4
|
Cas scaffolding protein family member 4 |
| chr7_-_126399778 | 0.10 |
ENSMUST00000141355.4
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr19_+_41818409 | 0.10 |
ENSMUST00000087155.5
|
Frat1
|
frequently rearranged in advanced T cell lymphomas |
| chr14_+_47069582 | 0.09 |
ENSMUST00000068532.10
|
Cgrrf1
|
cell growth regulator with ring finger domain 1 |
| chr6_+_134012602 | 0.09 |
ENSMUST00000081028.13
ENSMUST00000111963.8 |
Etv6
|
ets variant 6 |
| chr12_-_101943134 | 0.08 |
ENSMUST00000221227.2
|
Ndufb1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr12_-_72711509 | 0.08 |
ENSMUST00000221750.2
|
Dhrs7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr5_-_8417982 | 0.08 |
ENSMUST00000088761.11
ENSMUST00000115386.8 ENSMUST00000050166.14 ENSMUST00000046838.14 ENSMUST00000115388.9 ENSMUST00000088744.12 ENSMUST00000115385.2 |
Adam22
|
a disintegrin and metallopeptidase domain 22 |
| chr8_-_70980584 | 0.08 |
ENSMUST00000138260.8
ENSMUST00000117580.8 |
Kxd1
|
KxDL motif containing 1 |
| chr12_-_72711533 | 0.08 |
ENSMUST00000021512.11
|
Dhrs7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr10_+_81699768 | 0.08 |
ENSMUST00000220314.2
|
Gm32687
|
predicted gene, 32687 |
| chr11_-_69872050 | 0.08 |
ENSMUST00000108594.8
|
Elp5
|
elongator acetyltransferase complex subunit 5 |
| chr17_+_45817750 | 0.07 |
ENSMUST00000024733.9
|
Aars2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr7_-_30810422 | 0.07 |
ENSMUST00000039435.15
|
Hpn
|
hepsin |
| chrX_-_77627486 | 0.07 |
ENSMUST00000114025.8
ENSMUST00000134602.8 ENSMUST00000114024.9 |
Prrg1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr15_-_78456898 | 0.07 |
ENSMUST00000043214.8
|
Rac2
|
Rac family small GTPase 2 |
| chr14_-_52553764 | 0.06 |
ENSMUST00000135523.5
|
Sall2
|
spalt like transcription factor 2 |
| chr4_+_42922253 | 0.06 |
ENSMUST00000139100.2
|
Phf24
|
PHD finger protein 24 |
| chr2_+_134627987 | 0.06 |
ENSMUST00000131552.5
ENSMUST00000110116.8 |
Plcb1
|
phospholipase C, beta 1 |
| chr4_-_57300361 | 0.06 |
ENSMUST00000153926.8
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr14_+_47069667 | 0.06 |
ENSMUST00000140114.3
ENSMUST00000133989.8 |
Cgrrf1
|
cell growth regulator with ring finger domain 1 |
| chr4_+_126156118 | 0.06 |
ENSMUST00000030660.9
|
Trappc3
|
trafficking protein particle complex 3 |
| chr8_+_70980374 | 0.06 |
ENSMUST00000119353.9
ENSMUST00000075491.14 ENSMUST00000119698.8 |
Fkbp8
|
FK506 binding protein 8 |
| chr11_-_120348762 | 0.06 |
ENSMUST00000137632.2
ENSMUST00000044007.3 |
Oxld1
|
oxidoreductase like domain containing 1 |
| chr11_+_78403019 | 0.06 |
ENSMUST00000001127.11
|
Poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr2_+_118428690 | 0.06 |
ENSMUST00000038341.8
|
Bub1b
|
BUB1B, mitotic checkpoint serine/threonine kinase |
| chr12_-_79054050 | 0.06 |
ENSMUST00000056660.13
ENSMUST00000174721.8 |
Tmem229b
|
transmembrane protein 229B |
| chr4_+_101276474 | 0.06 |
ENSMUST00000102780.8
ENSMUST00000106946.8 ENSMUST00000106945.8 |
Ak4
|
adenylate kinase 4 |
| chr3_+_62245765 | 0.05 |
ENSMUST00000079300.13
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr11_+_100750177 | 0.05 |
ENSMUST00000004145.14
ENSMUST00000133036.8 |
Stat5a
|
signal transducer and activator of transcription 5A |
| chr2_-_167032068 | 0.05 |
ENSMUST00000059826.10
|
Kcnb1
|
potassium voltage gated channel, Shab-related subfamily, member 1 |
| chr8_+_4303067 | 0.05 |
ENSMUST00000011981.5
ENSMUST00000208316.2 |
Snapc2
|
small nuclear RNA activating complex, polypeptide 2 |
| chr4_-_134642286 | 0.05 |
ENSMUST00000105863.2
ENSMUST00000030626.12 |
Tmem50a
|
transmembrane protein 50A |
| chr10_+_79889322 | 0.05 |
ENSMUST00000105372.9
|
Gpx4
|
glutathione peroxidase 4 |
| chr11_+_69871952 | 0.05 |
ENSMUST00000108593.8
|
Ctdnep1
|
CTD nuclear envelope phosphatase 1 |
| chr2_+_90507552 | 0.05 |
ENSMUST00000057481.7
|
Nup160
|
nucleoporin 160 |
| chr13_-_56283331 | 0.05 |
ENSMUST00000045788.9
ENSMUST00000016081.13 |
Macroh2a1
|
macroH2A.1 histone |
| chr12_+_84332006 | 0.04 |
ENSMUST00000123614.8
ENSMUST00000147363.8 ENSMUST00000135001.8 ENSMUST00000146377.8 |
Ptgr2
|
prostaglandin reductase 2 |
| chr11_-_78402931 | 0.04 |
ENSMUST00000052566.8
|
Tmem199
|
transmembrane protein 199 |
| chr12_+_111540920 | 0.04 |
ENSMUST00000075281.8
ENSMUST00000084953.13 |
Mark3
|
MAP/microtubule affinity regulating kinase 3 |
| chr5_+_73071607 | 0.04 |
ENSMUST00000144843.8
|
Slain2
|
SLAIN motif family, member 2 |
| chr19_+_43678109 | 0.04 |
ENSMUST00000081079.6
|
Entpd7
|
ectonucleoside triphosphate diphosphohydrolase 7 |
| chr1_-_170978157 | 0.04 |
ENSMUST00000155798.2
ENSMUST00000081560.5 ENSMUST00000111336.10 |
Sdhc
|
succinate dehydrogenase complex, subunit C, integral membrane protein |
| chr10_+_81919146 | 0.04 |
ENSMUST00000220287.2
|
BC024063
|
cDNA sequence BC024063 |
| chr19_-_53932867 | 0.04 |
ENSMUST00000235688.2
ENSMUST00000235348.2 |
Bbip1
|
BBSome interacting protein 1 |
| chr2_-_106804679 | 0.04 |
ENSMUST00000028536.13
|
Arl14ep
|
ADP-ribosylation factor-like 14 effector protein |
| chr2_-_104241358 | 0.04 |
ENSMUST00000230671.2
|
D430041D05Rik
|
RIKEN cDNA D430041D05 gene |
| chr2_+_118730823 | 0.04 |
ENSMUST00000151162.2
|
Bahd1
|
bromo adjacent homology domain containing 1 |
| chr8_-_72178340 | 0.03 |
ENSMUST00000153800.8
ENSMUST00000146100.8 |
Fcho1
|
FCH domain only 1 |
| chr4_+_20042045 | 0.03 |
ENSMUST00000098242.4
|
Ggh
|
gamma-glutamyl hydrolase |
| chrX_+_20736405 | 0.03 |
ENSMUST00000115342.10
ENSMUST00000009530.5 |
Timp1
|
tissue inhibitor of metalloproteinase 1 |
| chr19_+_5138562 | 0.03 |
ENSMUST00000238093.2
ENSMUST00000025811.6 ENSMUST00000237025.2 |
Yif1a
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr6_-_122317484 | 0.03 |
ENSMUST00000112600.9
|
Phc1
|
polyhomeotic 1 |
| chr19_+_28941292 | 0.03 |
ENSMUST00000045674.4
|
Plpp6
|
phospholipid phosphatase 6 |
| chr13_-_43713583 | 0.03 |
ENSMUST00000021800.6
|
Mcur1
|
mitochondrial calcium uniporter regulator 1 |
| chr6_+_83055581 | 0.03 |
ENSMUST00000177177.8
ENSMUST00000176089.2 |
Pcgf1
|
polycomb group ring finger 1 |
| chr19_+_12775938 | 0.03 |
ENSMUST00000025601.8
|
Lpxn
|
leupaxin |
| chrX_-_103244784 | 0.03 |
ENSMUST00000118314.8
|
Nexmif
|
neurite extension and migration factor |
| chr7_+_15909010 | 0.03 |
ENSMUST00000176342.8
ENSMUST00000177540.8 |
Meis3
|
Meis homeobox 3 |
| chr18_+_31742565 | 0.02 |
ENSMUST00000164667.2
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene |
| chr4_-_42168603 | 0.02 |
ENSMUST00000098121.4
|
Gm13305
|
predicted gene 13305 |
| chr11_+_28803188 | 0.02 |
ENSMUST00000020759.12
|
Efemp1
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 1 |
| chr2_+_131048998 | 0.02 |
ENSMUST00000153097.3
|
Ap5s1
|
adaptor-related protein 5 complex, sigma 1 subunit |
| chr3_+_85946145 | 0.02 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr7_-_28661751 | 0.02 |
ENSMUST00000068045.14
ENSMUST00000217157.2 |
Actn4
|
actinin alpha 4 |
| chr15_-_82505132 | 0.02 |
ENSMUST00000109515.3
|
Cyp2d34
|
cytochrome P450, family 2, subfamily d, polypeptide 34 |
| chr1_-_180524587 | 0.02 |
ENSMUST00000027778.8
|
Mixl1
|
Mix1 homeobox-like 1 (Xenopus laevis) |
| chr7_-_44711130 | 0.02 |
ENSMUST00000211337.2
|
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chrX_+_158155171 | 0.02 |
ENSMUST00000087143.7
|
Eif1ax
|
eukaryotic translation initiation factor 1A, X-linked |
| chrX_-_103244703 | 0.02 |
ENSMUST00000087879.11
|
Nexmif
|
neurite extension and migration factor |
| chr4_-_140805613 | 0.02 |
ENSMUST00000030760.15
|
Necap2
|
NECAP endocytosis associated 2 |
| chr14_+_34395845 | 0.02 |
ENSMUST00000048263.14
|
Wapl
|
WAPL cohesin release factor |
| chr1_-_93563201 | 0.02 |
ENSMUST00000133769.8
|
Stk25
|
serine/threonine kinase 25 (yeast) |
| chr9_+_107457316 | 0.02 |
ENSMUST00000093785.6
|
Naa80
|
N(alpha)-acetyltransferase 80, NatH catalytic subunit |
| chr10_-_38998272 | 0.02 |
ENSMUST00000136546.8
|
Fam229b
|
family with sequence similarity 229, member B |
| chr15_+_82336535 | 0.02 |
ENSMUST00000089129.7
ENSMUST00000229313.2 ENSMUST00000231136.2 |
Cyp2d9
|
cytochrome P450, family 2, subfamily d, polypeptide 9 |
| chrX_-_163763273 | 0.02 |
ENSMUST00000112247.9
ENSMUST00000004715.2 |
Mospd2
|
motile sperm domain containing 2 |
| chrX_-_48886577 | 0.02 |
ENSMUST00000033442.14
ENSMUST00000114891.2 |
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr8_+_47193275 | 0.02 |
ENSMUST00000207571.3
|
Irf2
|
interferon regulatory factor 2 |
| chr7_+_111825063 | 0.02 |
ENSMUST00000050149.12
|
Mical2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr19_-_53932581 | 0.02 |
ENSMUST00000236885.2
ENSMUST00000236098.2 ENSMUST00000236370.2 |
Bbip1
|
BBSome interacting protein 1 |
| chr5_-_22549688 | 0.02 |
ENSMUST00000062372.14
ENSMUST00000161356.8 |
Reln
|
reelin |
| chrX_+_55917903 | 0.02 |
ENSMUST00000153784.8
|
Adgrg4
|
adhesion G protein-coupled receptor G4 |
| chr3_+_94280101 | 0.02 |
ENSMUST00000029795.10
|
Rorc
|
RAR-related orphan receptor gamma |
| chr10_-_77647677 | 0.02 |
ENSMUST00000095491.5
ENSMUST00000180157.2 |
Gm9507
|
predicted gene 9507 |
| chr10_+_77697785 | 0.01 |
ENSMUST00000218443.2
|
Gm3285
|
predicted gene 3285 |
| chr4_-_42665763 | 0.01 |
ENSMUST00000238770.2
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr11_+_23234644 | 0.01 |
ENSMUST00000150750.3
|
Xpo1
|
exportin 1 |
| chr5_+_73071695 | 0.01 |
ENSMUST00000143829.5
|
Slain2
|
SLAIN motif family, member 2 |
| chr9_+_54441404 | 0.01 |
ENSMUST00000118413.3
|
Sh2d7
|
SH2 domain containing 7 |
| chr10_+_77547249 | 0.01 |
ENSMUST00000075421.7
|
Gm10024
|
predicted gene 10024 |
| chr2_+_153334710 | 0.01 |
ENSMUST00000109783.2
|
4930404H24Rik
|
RIKEN cDNA 4930404H24 gene |
| chr11_-_72380730 | 0.01 |
ENSMUST00000045303.10
|
Spns2
|
spinster homolog 2 |
| chr8_-_70980293 | 0.01 |
ENSMUST00000132867.2
|
Kxd1
|
KxDL motif containing 1 |
| chr4_-_57300750 | 0.01 |
ENSMUST00000151964.2
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr6_-_144993451 | 0.01 |
ENSMUST00000123930.8
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr6_-_57956287 | 0.01 |
ENSMUST00000228585.2
|
Vmn1r25
|
vomeronasal 1 receptor 25 |
| chr10_-_77607159 | 0.01 |
ENSMUST00000168298.2
|
Gm3238
|
predicted gene 3238 |
| chrX_-_77627388 | 0.01 |
ENSMUST00000177904.8
|
Prrg1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr17_-_54153367 | 0.01 |
ENSMUST00000023886.7
|
Sult1c2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr3_-_145813802 | 0.01 |
ENSMUST00000160285.2
|
Dnai3
|
dynein axonemal intermediate chain 3 |
| chr7_+_119289249 | 0.01 |
ENSMUST00000047045.10
|
Acsm4
|
acyl-CoA synthetase medium-chain family member 4 |
| chr7_-_44711075 | 0.01 |
ENSMUST00000007981.9
ENSMUST00000210500.2 ENSMUST00000210493.2 |
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr17_+_21506795 | 0.01 |
ENSMUST00000066998.5
|
Vmn1r236
|
vomeronasal 1 receptor 236 |
| chr10_-_79624758 | 0.01 |
ENSMUST00000020573.13
|
Prss57
|
protease, serine 57 |
| chr2_-_127571836 | 0.01 |
ENSMUST00000028856.3
|
Mall
|
mal, T cell differentiation protein-like |
| chr4_-_97472844 | 0.01 |
ENSMUST00000107067.8
ENSMUST00000107068.9 |
E130114P18Rik
|
RIKEN cDNA E130114P18 gene |
| chr10_-_77624034 | 0.01 |
ENSMUST00000180161.2
|
Gm7137
|
predicted gene 7137 |
| chr15_-_79048674 | 0.01 |
ENSMUST00000230261.2
ENSMUST00000040019.5 |
Sox10
|
SRY (sex determining region Y)-box 10 |
| chr11_+_53992054 | 0.01 |
ENSMUST00000135653.8
|
P4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha II polypeptide |
| chr5_+_31274064 | 0.01 |
ENSMUST00000202769.2
|
Trim54
|
tripartite motif-containing 54 |
| chr5_+_73071897 | 0.01 |
ENSMUST00000200785.2
|
Slain2
|
SLAIN motif family, member 2 |
| chr6_+_90246088 | 0.01 |
ENSMUST00000058039.3
|
Vmn1r54
|
vomeronasal 1 receptor 54 |
| chr5_+_31274046 | 0.01 |
ENSMUST00000013771.15
|
Trim54
|
tripartite motif-containing 54 |
| chr10_+_77688644 | 0.01 |
ENSMUST00000186874.3
ENSMUST00000217996.2 |
Gm10318
|
predicted gene 10318 |
| chr9_-_106448182 | 0.01 |
ENSMUST00000085111.5
|
Iqcf4
|
IQ motif containing F4 |
| chr8_+_47192911 | 0.01 |
ENSMUST00000208433.2
|
Irf2
|
interferon regulatory factor 2 |
| chr5_+_92135294 | 0.01 |
ENSMUST00000178614.3
|
Odaph
|
odontogenesis associated phosphoprotein |
| chr18_-_61147272 | 0.01 |
ENSMUST00000025520.10
|
Slc6a7
|
solute carrier family 6 (neurotransmitter transporter, L-proline), member 7 |
| chrX_-_103244728 | 0.01 |
ENSMUST00000056502.7
|
Nexmif
|
neurite extension and migration factor |
| chr2_+_160722562 | 0.01 |
ENSMUST00000109456.9
|
Lpin3
|
lipin 3 |
| chr6_+_49372463 | 0.01 |
ENSMUST00000024171.14
ENSMUST00000163954.8 ENSMUST00000172459.8 |
Stk31
|
serine threonine kinase 31 |
| chr18_+_61238908 | 0.01 |
ENSMUST00000115268.4
|
Csf1r
|
colony stimulating factor 1 receptor |
| chr15_+_16778187 | 0.01 |
ENSMUST00000026432.8
|
Cdh9
|
cadherin 9 |
| chr12_+_111387023 | 0.01 |
ENSMUST00000220852.2
|
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr14_-_21791544 | 0.01 |
ENSMUST00000184703.8
ENSMUST00000183698.8 ENSMUST00000119866.9 |
Dusp13
|
dual specificity phosphatase 13 |
| chr15_+_82439273 | 0.01 |
ENSMUST00000229103.2
ENSMUST00000068861.8 ENSMUST00000229904.2 |
Cyp2d12
|
cytochrome P450, family 2, subfamily d, polypeptide 12 |
| chr10_-_81121596 | 0.01 |
ENSMUST00000218484.2
|
Tjp3
|
tight junction protein 3 |
| chr12_+_88327607 | 0.01 |
ENSMUST00000166940.3
|
Adck1
|
aarF domain containing kinase 1 |
| chr9_-_106533279 | 0.01 |
ENSMUST00000023959.13
ENSMUST00000201681.2 |
Grm2
|
glutamate receptor, metabotropic 2 |
| chr11_-_47270201 | 0.01 |
ENSMUST00000077221.6
|
Sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr7_-_46854668 | 0.01 |
ENSMUST00000186456.2
|
Mrgpra6
|
MAS-related GPR, member A6 |
| chr11_+_42310557 | 0.01 |
ENSMUST00000007797.10
|
Gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 2 |
| chr10_+_80134917 | 0.01 |
ENSMUST00000154212.8
|
Apc2
|
APC regulator of WNT signaling pathway 2 |
| chr4_+_103476777 | 0.01 |
ENSMUST00000106827.8
|
Dab1
|
disabled 1 |
| chr2_-_79959802 | 0.01 |
ENSMUST00000102653.8
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr1_+_50966670 | 0.01 |
ENSMUST00000081851.4
|
Tmeff2
|
transmembrane protein with EGF-like and two follistatin-like domains 2 |
| chr10_+_90665639 | 0.01 |
ENSMUST00000179337.9
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr10_+_90665399 | 0.00 |
ENSMUST00000179694.9
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr6_-_126717590 | 0.00 |
ENSMUST00000185333.2
|
Kcna6
|
potassium voltage-gated channel, shaker-related, subfamily, member 6 |
| chr6_-_83513222 | 0.00 |
ENSMUST00000075161.12
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr10_+_90665270 | 0.00 |
ENSMUST00000182202.8
ENSMUST00000182966.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr4_-_139560229 | 0.00 |
ENSMUST00000174681.2
|
Pax7
|
paired box 7 |
| chr14_+_53707213 | 0.00 |
ENSMUST00000103643.4
|
Trav8-1
|
T cell receptor alpha variable 8-1 |
| chr10_-_77662926 | 0.00 |
ENSMUST00000092369.4
|
Krtap10-4
|
keratin associated protein 10-4 |
| chr6_-_57367651 | 0.00 |
ENSMUST00000164732.2
|
Vmn1r18
|
vomeronasal 1 receptor 18 |
| chr6_-_57338363 | 0.00 |
ENSMUST00000227966.2
|
Vmn1r17
|
vomeronasal 1 receptor 17 |
| chr5_-_131567526 | 0.00 |
ENSMUST00000161374.8
|
Auts2
|
autism susceptibility candidate 2 |
| chr6_-_126717114 | 0.00 |
ENSMUST00000112242.2
|
Kcna6
|
potassium voltage-gated channel, shaker-related, subfamily, member 6 |
| chr15_-_82291372 | 0.00 |
ENSMUST00000230198.2
ENSMUST00000230248.2 ENSMUST00000072776.5 ENSMUST00000229911.2 |
Cyp2d10
|
cytochrome P450, family 2, subfamily d, polypeptide 10 |
| chr15_+_83664196 | 0.00 |
ENSMUST00000046168.12
|
Mpped1
|
metallophosphoesterase domain containing 1 |
| chr2_-_121381832 | 0.00 |
ENSMUST00000212518.2
|
Frmd5
|
FERM domain containing 5 |
| chr11_-_61344818 | 0.00 |
ENSMUST00000060255.14
ENSMUST00000054927.14 ENSMUST00000102661.4 |
Rnf112
|
ring finger protein 112 |
| chr1_-_93563406 | 0.00 |
ENSMUST00000027498.14
|
Stk25
|
serine/threonine kinase 25 (yeast) |
| chr15_-_82278223 | 0.00 |
ENSMUST00000170255.2
|
Cyp2d11
|
cytochrome P450, family 2, subfamily d, polypeptide 11 |
| chr12_-_17061504 | 0.00 |
ENSMUST00000189479.2
|
2410004P03Rik
|
RIKEN cDNA 2410004P03 gene |
| chr18_-_52662917 | 0.00 |
ENSMUST00000171470.8
|
Lox
|
lysyl oxidase |
| chr6_-_144993362 | 0.00 |
ENSMUST00000149769.6
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr2_-_27138347 | 0.00 |
ENSMUST00000139312.8
|
Sardh
|
sarcosine dehydrogenase |
| chr4_-_60222580 | 0.00 |
ENSMUST00000095058.5
ENSMUST00000163931.8 |
Mup8
|
major urinary protein 8 |
| chr4_+_131648509 | 0.00 |
ENSMUST00000238733.2
|
Tmem200b
|
transmembrane protein 200B |
| chr12_-_87283780 | 0.00 |
ENSMUST00000221768.2
|
Noxred1
|
NADP+ dependent oxidoreductase domain containing 1 |
| chr8_+_34006758 | 0.00 |
ENSMUST00000149399.8
|
Tex15
|
testis expressed gene 15 |
| chr17_+_33418036 | 0.00 |
ENSMUST00000112165.4
|
Olfr239
|
olfactory receptor 239 |
| chr2_+_172314433 | 0.00 |
ENSMUST00000029007.3
|
Fam209
|
family with sequence similarity 209 |
| chr7_-_28661648 | 0.00 |
ENSMUST00000127210.8
|
Actn4
|
actinin alpha 4 |
| chr17_+_33395390 | 0.00 |
ENSMUST00000112168.4
|
Olfr55
|
olfactory receptor 55 |
| chr12_+_85157607 | 0.00 |
ENSMUST00000053811.10
|
Dlst
|
dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) |
| chr5_-_134668152 | 0.00 |
ENSMUST00000036125.10
ENSMUST00000202622.4 |
Eif4h
|
eukaryotic translation initiation factor 4H |
| chr11_+_104468107 | 0.00 |
ENSMUST00000106956.10
|
Myl4
|
myosin, light polypeptide 4 |
| chr6_-_99703295 | 0.00 |
ENSMUST00000032152.14
|
Prok2
|
prokineticin 2 |
| chr16_+_90182895 | 0.00 |
ENSMUST00000065856.8
|
Hunk
|
hormonally upregulated Neu-associated kinase |
| chr6_+_83055321 | 0.00 |
ENSMUST00000165164.9
ENSMUST00000092614.9 |
Pcgf1
|
polycomb group ring finger 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zkscan1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.2 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.2 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.2 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.0 | 0.1 | GO:1904117 | interleukin-12-mediated signaling pathway(GO:0035722) activation of meiosis involved in egg activation(GO:0060466) cellular response to interleukin-12(GO:0071349) response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.3 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.0 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 0.2 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.4 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.0 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.0 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |