Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
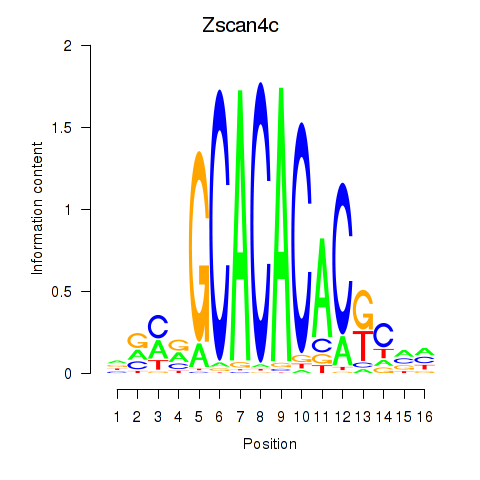
Results for Zscan4c
Z-value: 0.70
Transcription factors associated with Zscan4c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zscan4c
|
ENSMUSG00000054272.7 | zinc finger and SCAN domain containing 4C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zscan4c | mm39_v1_chr7_+_10739672_10739672 | -0.73 | 1.6e-01 | Click! |
Activity profile of Zscan4c motif
Sorted Z-values of Zscan4c motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_61431271 | 0.62 |
ENSMUST00000020287.8
|
Npffr1
|
neuropeptide FF receptor 1 |
| chr4_-_87724533 | 0.53 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr18_-_42395207 | 0.48 |
ENSMUST00000097590.5
|
Lars
|
leucyl-tRNA synthetase |
| chr4_-_87724512 | 0.44 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr7_+_46490899 | 0.38 |
ENSMUST00000147535.8
|
Ldha
|
lactate dehydrogenase A |
| chr10_-_37014859 | 0.38 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr11_-_101676076 | 0.37 |
ENSMUST00000164750.8
ENSMUST00000107176.8 ENSMUST00000017868.7 |
Etv4
|
ets variant 4 |
| chr4_-_136620376 | 0.34 |
ENSMUST00000046332.6
|
C1qc
|
complement component 1, q subcomponent, C chain |
| chr19_-_6117815 | 0.33 |
ENSMUST00000162575.8
ENSMUST00000159084.8 ENSMUST00000161718.8 ENSMUST00000162810.8 ENSMUST00000025713.12 ENSMUST00000113543.9 ENSMUST00000160417.8 ENSMUST00000161528.2 |
Tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr15_-_77037972 | 0.32 |
ENSMUST00000111581.4
ENSMUST00000166610.8 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr15_+_44482944 | 0.30 |
ENSMUST00000022964.9
|
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr7_-_132723918 | 0.26 |
ENSMUST00000172341.8
|
Ctbp2
|
C-terminal binding protein 2 |
| chr18_-_42395131 | 0.25 |
ENSMUST00000236102.2
|
Lars
|
leucyl-tRNA synthetase |
| chr7_-_108774367 | 0.24 |
ENSMUST00000207178.2
|
Lmo1
|
LIM domain only 1 |
| chr13_-_9815173 | 0.23 |
ENSMUST00000062658.15
ENSMUST00000222358.2 |
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr13_-_9815350 | 0.22 |
ENSMUST00000110636.9
ENSMUST00000152725.8 |
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr12_+_52550775 | 0.22 |
ENSMUST00000219443.2
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr2_+_30254239 | 0.20 |
ENSMUST00000077977.14
ENSMUST00000140075.9 ENSMUST00000142801.8 ENSMUST00000100214.10 |
Miga2
|
mitoguardin 2 |
| chr13_-_9814979 | 0.19 |
ENSMUST00000110634.8
|
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr3_-_107667499 | 0.19 |
ENSMUST00000153114.2
ENSMUST00000118593.8 ENSMUST00000120243.8 |
Csf1
|
colony stimulating factor 1 (macrophage) |
| chr11_-_70128712 | 0.17 |
ENSMUST00000108577.8
ENSMUST00000108579.8 ENSMUST00000021181.13 ENSMUST00000108578.9 ENSMUST00000102569.10 |
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr15_+_44482545 | 0.17 |
ENSMUST00000227691.2
|
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr13_-_9815276 | 0.17 |
ENSMUST00000130151.8
|
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr2_-_157988303 | 0.17 |
ENSMUST00000103122.10
|
Tgm2
|
transglutaminase 2, C polypeptide |
| chr11_-_70128462 | 0.17 |
ENSMUST00000100950.10
|
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr4_-_129534403 | 0.15 |
ENSMUST00000084264.12
|
Txlna
|
taxilin alpha |
| chr8_-_117459730 | 0.15 |
ENSMUST00000109102.4
|
Cdyl2
|
chromodomain protein, Y chromosome-like 2 |
| chr10_+_18345706 | 0.15 |
ENSMUST00000162891.8
ENSMUST00000100054.4 |
Nhsl1
|
NHS-like 1 |
| chr15_-_77037756 | 0.15 |
ENSMUST00000227314.2
ENSMUST00000227930.2 ENSMUST00000227533.2 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr3_-_51184730 | 0.15 |
ENSMUST00000195432.2
ENSMUST00000091144.11 ENSMUST00000156983.3 |
Elf2
|
E74-like factor 2 |
| chr13_-_9814901 | 0.15 |
ENSMUST00000223421.2
ENSMUST00000128658.8 |
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr15_+_44482667 | 0.14 |
ENSMUST00000228648.2
ENSMUST00000226165.2 |
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr16_-_78373537 | 0.14 |
ENSMUST00000232052.2
ENSMUST00000114219.8 ENSMUST00000114218.2 |
D16Ertd472e
|
DNA segment, Chr 16, ERATO Doi 472, expressed |
| chr12_+_52551092 | 0.14 |
ENSMUST00000217820.2
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr16_-_78373510 | 0.14 |
ENSMUST00000231973.2
ENSMUST00000232528.2 ENSMUST00000114220.9 |
D16Ertd472e
|
DNA segment, Chr 16, ERATO Doi 472, expressed |
| chr5_+_115034997 | 0.13 |
ENSMUST00000031542.13
ENSMUST00000146072.8 ENSMUST00000150361.2 |
Oasl2
|
2'-5' oligoadenylate synthetase-like 2 |
| chr15_-_77037926 | 0.13 |
ENSMUST00000228087.2
|
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr5_-_96309600 | 0.12 |
ENSMUST00000129646.8
ENSMUST00000113005.9 ENSMUST00000154500.2 ENSMUST00000141383.8 |
Cnot6l
|
CCR4-NOT transcription complex, subunit 6-like |
| chr3_-_51184895 | 0.12 |
ENSMUST00000108051.8
ENSMUST00000108053.9 |
Elf2
|
E74-like factor 2 |
| chr9_+_21279802 | 0.12 |
ENSMUST00000214474.2
|
Ilf3
|
interleukin enhancer binding factor 3 |
| chr13_-_76205115 | 0.11 |
ENSMUST00000056130.8
|
Gpr150
|
G protein-coupled receptor 150 |
| chr5_-_145138639 | 0.10 |
ENSMUST00000162220.2
ENSMUST00000031632.9 ENSMUST00000198959.2 |
Zkscan14
|
zinc finger with KRAB and SCAN domains 14 |
| chr16_-_78373570 | 0.10 |
ENSMUST00000231272.2
|
D16Ertd472e
|
DNA segment, Chr 16, ERATO Doi 472, expressed |
| chr1_+_87983099 | 0.10 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr9_+_111011388 | 0.10 |
ENSMUST00000217117.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr1_+_87983189 | 0.09 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr11_-_109364046 | 0.09 |
ENSMUST00000070872.13
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr6_+_48570817 | 0.08 |
ENSMUST00000154010.8
ENSMUST00000009420.15 ENSMUST00000163452.7 ENSMUST00000118229.2 ENSMUST00000135151.3 |
Repin1
|
replication initiator 1 |
| chr19_-_46033353 | 0.08 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr9_+_21848282 | 0.08 |
ENSMUST00000046371.13
|
Plppr2
|
phospholipid phosphatase related 2 |
| chr11_-_70128587 | 0.08 |
ENSMUST00000108576.10
|
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr9_-_50639367 | 0.08 |
ENSMUST00000117646.8
|
Dixdc1
|
DIX domain containing 1 |
| chr6_-_145811274 | 0.08 |
ENSMUST00000032386.11
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr11_-_70128678 | 0.07 |
ENSMUST00000108575.9
|
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr3_-_135313982 | 0.06 |
ENSMUST00000132668.8
|
Nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
| chr19_+_53298906 | 0.06 |
ENSMUST00000003870.15
|
Mxi1
|
MAX interactor 1, dimerization protein |
| chr19_-_28657477 | 0.05 |
ENSMUST00000162022.8
ENSMUST00000112612.9 |
Glis3
|
GLIS family zinc finger 3 |
| chr11_-_113642135 | 0.05 |
ENSMUST00000106616.2
|
Cdc42ep4
|
CDC42 effector protein (Rho GTPase binding) 4 |
| chr11_-_113641980 | 0.05 |
ENSMUST00000153453.2
|
Cdc42ep4
|
CDC42 effector protein (Rho GTPase binding) 4 |
| chr13_+_55517545 | 0.05 |
ENSMUST00000063771.14
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr9_+_111011327 | 0.05 |
ENSMUST00000216430.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr4_+_114914880 | 0.05 |
ENSMUST00000161601.8
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr9_+_37466989 | 0.04 |
ENSMUST00000213126.2
|
Siae
|
sialic acid acetylesterase |
| chr10_-_81360059 | 0.04 |
ENSMUST00000043709.8
|
Gna15
|
guanine nucleotide binding protein, alpha 15 |
| chr15_-_98705791 | 0.03 |
ENSMUST00000075444.8
|
Ddn
|
dendrin |
| chr2_+_85804239 | 0.03 |
ENSMUST00000217244.2
|
Olfr1029
|
olfactory receptor 1029 |
| chr18_+_64473091 | 0.03 |
ENSMUST00000175965.10
|
Onecut2
|
one cut domain, family member 2 |
| chrX_+_47608122 | 0.02 |
ENSMUST00000033430.3
|
Rab33a
|
RAB33A, member RAS oncogene family |
| chr11_+_49442576 | 0.02 |
ENSMUST00000167248.3
|
Olfr1381
|
olfactory receptor 1381 |
| chr7_-_45519853 | 0.02 |
ENSMUST00000211713.2
|
Grin2d
|
glutamate receptor, ionotropic, NMDA2D (epsilon 4) |
| chr3_-_63758672 | 0.02 |
ENSMUST00000162269.9
ENSMUST00000159676.9 ENSMUST00000175947.8 |
Plch1
|
phospholipase C, eta 1 |
| chr11_-_113642635 | 0.02 |
ENSMUST00000053536.5
|
Cdc42ep4
|
CDC42 effector protein (Rho GTPase binding) 4 |
| chr6_-_145811028 | 0.02 |
ENSMUST00000111703.2
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr10_+_87694117 | 0.02 |
ENSMUST00000122386.8
|
Igf1
|
insulin-like growth factor 1 |
| chr11_+_49426014 | 0.01 |
ENSMUST00000074543.2
|
Olfr1382
|
olfactory receptor 1382 |
| chr3_+_133942244 | 0.01 |
ENSMUST00000181904.3
|
Cxxc4
|
CXXC finger 4 |
| chr9_+_43978369 | 0.01 |
ENSMUST00000177054.8
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr15_-_37458768 | 0.01 |
ENSMUST00000116445.9
|
Ncald
|
neurocalcin delta |
| chr19_-_21449916 | 0.01 |
ENSMUST00000087600.10
|
Gda
|
guanine deaminase |
| chr2_-_35994819 | 0.01 |
ENSMUST00000148852.4
|
Lhx6
|
LIM homeobox protein 6 |
| chr5_-_129030111 | 0.01 |
ENSMUST00000196085.5
|
Rimbp2
|
RIMS binding protein 2 |
| chr5_-_44956932 | 0.01 |
ENSMUST00000199261.2
ENSMUST00000199534.5 |
Ldb2
|
LIM domain binding 2 |
| chr14_+_122712809 | 0.01 |
ENSMUST00000075888.6
|
Zic2
|
zinc finger protein of the cerebellum 2 |
| chr6_+_91661074 | 0.01 |
ENSMUST00000205480.2
ENSMUST00000206545.2 |
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chr9_+_26645024 | 0.01 |
ENSMUST00000160899.8
ENSMUST00000161431.3 ENSMUST00000159799.8 |
B3gat1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) |
| chr6_+_52290415 | 0.01 |
ENSMUST00000031787.8
ENSMUST00000129243.3 |
Evx1
|
even-skipped homeobox 1 |
| chr13_-_114595122 | 0.01 |
ENSMUST00000231252.2
|
Fst
|
follistatin |
| chr3_+_125197722 | 0.01 |
ENSMUST00000173932.8
|
Ndst4
|
N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 |
| chr10_-_114638202 | 0.00 |
ENSMUST00000239411.2
|
Trhde
|
TRH-degrading enzyme |
| chr5_-_44956981 | 0.00 |
ENSMUST00000070748.10
|
Ldb2
|
LIM domain binding 2 |
| chr10_+_102348076 | 0.00 |
ENSMUST00000219445.2
|
Rassf9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr16_-_9812410 | 0.00 |
ENSMUST00000115835.8
|
Grin2a
|
glutamate receptor, ionotropic, NMDA2A (epsilon 1) |
| chr5_-_44957016 | 0.00 |
ENSMUST00000199256.5
|
Ldb2
|
LIM domain binding 2 |
| chr11_-_37126709 | 0.00 |
ENSMUST00000102801.8
|
Tenm2
|
teneurin transmembrane protein 2 |
| chr14_+_117163215 | 0.00 |
ENSMUST00000078849.12
|
Gpc6
|
glypican 6 |
| chrX_-_160942713 | 0.00 |
ENSMUST00000087085.10
|
Nhs
|
NHS actin remodeling regulator |
| chr10_+_84938452 | 0.00 |
ENSMUST00000095383.6
|
Tmem263
|
transmembrane protein 263 |
| chr16_-_38533597 | 0.00 |
ENSMUST00000023487.5
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr9_+_26645141 | 0.00 |
ENSMUST00000115269.9
|
B3gat1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) |
| chr3_+_13538266 | 0.00 |
ENSMUST00000211860.2
|
Ralyl
|
RALY RNA binding protein-like |
| chr7_+_50248918 | 0.00 |
ENSMUST00000119710.3
|
4933405O20Rik
|
RIKEN cDNA 4933405O20 gene |
| chr13_+_58955675 | 0.00 |
ENSMUST00000224402.2
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr11_-_52036355 | 0.00 |
ENSMUST00000078264.2
|
Olfr1373
|
olfactory receptor 1373 |
| chr9_-_31824758 | 0.00 |
ENSMUST00000116615.5
|
Barx2
|
BarH-like homeobox 2 |
| chr19_-_43879031 | 0.00 |
ENSMUST00000212048.2
|
Dnmbp
|
dynamin binding protein |
| chr2_-_115895528 | 0.00 |
ENSMUST00000028639.13
ENSMUST00000102538.11 |
Meis2
|
Meis homeobox 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zscan4c
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.1 | 0.2 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.1 | 0.2 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 1.0 | GO:2000096 | segment specification(GO:0007379) positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.6 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.3 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.4 | GO:0019659 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.4 | GO:0060762 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.0 | 0.1 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.0 | 1.0 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:1904632 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) response to diterpene(GO:1904629) cellular response to diterpene(GO:1904630) response to glucoside(GO:1904631) cellular response to glucoside(GO:1904632) |
| 0.0 | 0.2 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.0 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.2 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 1.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.0 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.0 | 0.3 | GO:0097470 | ribbon synapse(GO:0097470) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0004823 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 0.1 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0071820 | N-box binding(GO:0071820) |
| 0.0 | 0.3 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |