Project
avrg: 12D miR HR13_24
Navigation
Downloads
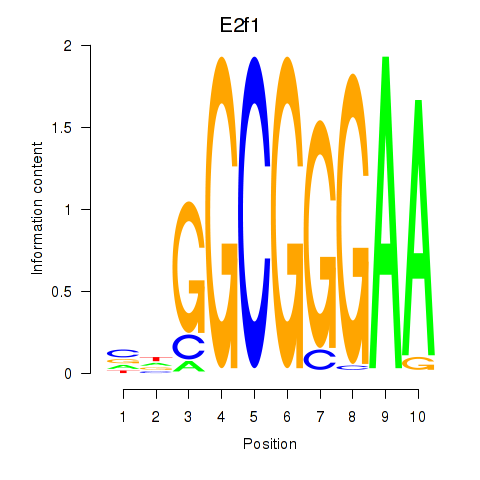
Results for E2f1
Z-value: 6.42
Transcription factors associated with E2f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f1
|
ENSMUSG00000027490.11 | E2F transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f1 | mm10_v2_chr2_-_154569845_154569892 | 0.91 | 1.1e-04 | Click! |
Activity profile of E2f1 motif
Sorted Z-values of E2f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_89294608 | 9.67 |
ENSMUST00000107131.1
|
Cdkn2a
|
cyclin-dependent kinase inhibitor 2A |
| chr4_+_126556935 | 8.33 |
ENSMUST00000048391.8
|
Clspn
|
claspin |
| chr1_-_20820213 | 7.89 |
ENSMUST00000053266.9
|
Mcm3
|
minichromosome maintenance deficient 3 (S. cerevisiae) |
| chr5_+_45669907 | 7.80 |
ENSMUST00000117396.1
|
Ncapg
|
non-SMC condensin I complex, subunit G |
| chr9_+_65890237 | 7.61 |
ENSMUST00000045802.6
|
2810417H13Rik
|
RIKEN cDNA 2810417H13 gene |
| chr5_+_76840597 | 7.12 |
ENSMUST00000120639.2
ENSMUST00000163347.1 ENSMUST00000121851.1 |
C530008M17Rik
|
RIKEN cDNA C530008M17 gene |
| chr17_+_56303396 | 6.83 |
ENSMUST00000113038.1
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr1_+_180641330 | 6.79 |
ENSMUST00000085804.5
|
Lin9
|
lin-9 homolog (C. elegans) |
| chr17_+_56303321 | 6.52 |
ENSMUST00000001258.8
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr13_-_55329723 | 6.31 |
ENSMUST00000021941.7
|
Mxd3
|
Max dimerization protein 3 |
| chr10_+_110745433 | 6.31 |
ENSMUST00000174857.1
ENSMUST00000073781.5 ENSMUST00000173471.1 ENSMUST00000173634.1 |
E2f7
|
E2F transcription factor 7 |
| chr7_+_102441685 | 6.17 |
ENSMUST00000033283.9
|
Rrm1
|
ribonucleotide reductase M1 |
| chr15_-_82212796 | 6.12 |
ENSMUST00000179269.1
|
AI848285
|
expressed sequence AI848285 |
| chr1_-_191575534 | 6.06 |
ENSMUST00000027933.5
|
Dtl
|
denticleless homolog (Drosophila) |
| chr14_-_47276790 | 5.73 |
ENSMUST00000111792.1
ENSMUST00000111791.1 ENSMUST00000111790.1 |
Wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr5_-_134747241 | 5.68 |
ENSMUST00000015138.9
|
Eln
|
elastin |
| chr6_+_134929118 | 5.55 |
ENSMUST00000185152.1
ENSMUST00000184504.1 |
RP23-45G16.5
|
RP23-45G16.5 |
| chr11_+_98907801 | 5.37 |
ENSMUST00000092706.6
|
Cdc6
|
cell division cycle 6 |
| chr17_+_35841668 | 5.32 |
ENSMUST00000174124.1
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr16_+_93883895 | 5.31 |
ENSMUST00000023666.4
ENSMUST00000117099.1 ENSMUST00000142316.1 |
Chaf1b
|
chromatin assembly factor 1, subunit B (p60) |
| chr13_-_100775844 | 5.20 |
ENSMUST00000075550.3
|
Cenph
|
centromere protein H |
| chr12_+_111271089 | 5.11 |
ENSMUST00000021707.6
|
Amn
|
amnionless |
| chr10_+_80356459 | 5.00 |
ENSMUST00000039836.8
ENSMUST00000105351.1 |
Plk5
|
polo-like kinase 5 |
| chr12_+_69168808 | 4.93 |
ENSMUST00000110621.1
|
Lrr1
|
leucine rich repeat protein 1 |
| chr12_+_116405397 | 4.91 |
ENSMUST00000084828.3
|
Ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr4_+_126556994 | 4.90 |
ENSMUST00000147675.1
|
Clspn
|
claspin |
| chr8_+_71406003 | 4.89 |
ENSMUST00000119976.1
ENSMUST00000120725.1 |
Ankle1
|
ankyrin repeat and LEM domain containing 1 |
| chr6_+_134929089 | 4.89 |
ENSMUST00000183867.1
ENSMUST00000184991.1 ENSMUST00000183905.1 |
RP23-45G16.5
|
RP23-45G16.5 |
| chr4_+_115000156 | 4.82 |
ENSMUST00000030490.6
|
Stil
|
Scl/Tal1 interrupting locus |
| chr5_+_123749696 | 4.77 |
ENSMUST00000031366.7
|
Kntc1
|
kinetochore associated 1 |
| chr9_+_106477269 | 4.66 |
ENSMUST00000047721.8
|
Rrp9
|
RRP9, small subunit (SSU) processome component, homolog (yeast) |
| chr10_+_110920170 | 4.66 |
ENSMUST00000020403.5
|
Csrp2
|
cysteine and glycine-rich protein 2 |
| chr8_+_46617426 | 4.63 |
ENSMUST00000093517.5
|
Casp3
|
caspase 3 |
| chr1_+_92831614 | 4.62 |
ENSMUST00000045970.6
|
Gpc1
|
glypican 1 |
| chr17_-_35516780 | 4.57 |
ENSMUST00000160885.1
ENSMUST00000159009.1 ENSMUST00000161012.1 |
Tcf19
|
transcription factor 19 |
| chr10_+_128232065 | 4.50 |
ENSMUST00000055539.4
ENSMUST00000105244.1 ENSMUST00000105243.2 ENSMUST00000125289.1 ENSMUST00000105242.1 |
Timeless
|
timeless circadian clock 1 |
| chr9_-_61946768 | 4.46 |
ENSMUST00000034815.7
|
Kif23
|
kinesin family member 23 |
| chrY_+_90784738 | 4.44 |
ENSMUST00000179483.1
|
Erdr1
|
erythroid differentiation regulator 1 |
| chr7_+_13278778 | 4.37 |
ENSMUST00000098814.4
ENSMUST00000146998.1 ENSMUST00000185145.1 |
Lig1
|
ligase I, DNA, ATP-dependent |
| chr4_+_115000174 | 4.34 |
ENSMUST00000129957.1
|
Stil
|
Scl/Tal1 interrupting locus |
| chr2_+_163054682 | 4.30 |
ENSMUST00000018005.3
|
Mybl2
|
myeloblastosis oncogene-like 2 |
| chr4_-_91372028 | 4.29 |
ENSMUST00000107110.1
ENSMUST00000008633.8 ENSMUST00000107118.1 |
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B) |
| chr5_+_76657673 | 4.20 |
ENSMUST00000128112.1
|
C530008M17Rik
|
RIKEN cDNA C530008M17 gene |
| chr16_-_18811615 | 4.19 |
ENSMUST00000096990.3
|
Cdc45
|
cell division cycle 45 |
| chr12_+_24708241 | 4.18 |
ENSMUST00000020980.5
|
Rrm2
|
ribonucleotide reductase M2 |
| chr15_-_78773452 | 4.18 |
ENSMUST00000018313.5
|
Mfng
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr6_+_49822710 | 4.17 |
ENSMUST00000031843.6
|
Npy
|
neuropeptide Y |
| chr15_-_58135047 | 4.17 |
ENSMUST00000038194.3
|
Atad2
|
ATPase family, AAA domain containing 2 |
| chr6_-_126939524 | 4.06 |
ENSMUST00000144954.1
ENSMUST00000112221.1 ENSMUST00000112220.1 |
Rad51ap1
|
RAD51 associated protein 1 |
| chr4_+_24496434 | 4.06 |
ENSMUST00000108222.2
ENSMUST00000138567.2 ENSMUST00000050446.6 |
Mms22l
|
MMS22-like, DNA repair protein |
| chr15_+_55557399 | 4.05 |
ENSMUST00000022998.7
|
Mtbp
|
Mdm2, transformed 3T3 cell double minute p53 binding protein |
| chrX_-_111463149 | 4.04 |
ENSMUST00000096348.3
ENSMUST00000113428.2 |
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr16_-_4559720 | 4.03 |
ENSMUST00000005862.7
|
Tfap4
|
transcription factor AP4 |
| chr17_+_87672523 | 4.02 |
ENSMUST00000172855.1
|
Msh2
|
mutS homolog 2 (E. coli) |
| chr8_+_75109528 | 4.00 |
ENSMUST00000164309.1
|
Mcm5
|
minichromosome maintenance deficient 5, cell division cycle 46 (S. cerevisiae) |
| chr5_-_138171248 | 3.99 |
ENSMUST00000153867.1
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr13_-_47105790 | 3.92 |
ENSMUST00000129352.1
|
Dek
|
DEK oncogene (DNA binding) |
| chr13_-_47106176 | 3.79 |
ENSMUST00000021807.6
ENSMUST00000135278.1 |
Dek
|
DEK oncogene (DNA binding) |
| chr4_+_52439235 | 3.74 |
ENSMUST00000117280.1
ENSMUST00000102915.3 ENSMUST00000142227.1 |
Smc2
|
structural maintenance of chromosomes 2 |
| chr6_+_113531675 | 3.74 |
ENSMUST00000036340.5
ENSMUST00000101051.2 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr4_+_128993224 | 3.68 |
ENSMUST00000030583.6
ENSMUST00000102604.4 |
Ak2
|
adenylate kinase 2 |
| chr12_+_117843873 | 3.67 |
ENSMUST00000176735.1
ENSMUST00000177339.1 |
Cdca7l
|
cell division cycle associated 7 like |
| chr13_-_112652295 | 3.66 |
ENSMUST00000099166.2
ENSMUST00000075748.5 |
Ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr4_-_137796350 | 3.66 |
ENSMUST00000030551.4
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr11_-_69948145 | 3.64 |
ENSMUST00000179298.1
ENSMUST00000018710.6 ENSMUST00000135437.1 ENSMUST00000141837.2 ENSMUST00000142500.1 |
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr19_-_9899450 | 3.58 |
ENSMUST00000025562.7
|
Incenp
|
inner centromere protein |
| chr16_+_37011758 | 3.57 |
ENSMUST00000071452.5
ENSMUST00000054034.6 |
Polq
|
polymerase (DNA directed), theta |
| chr1_+_172482199 | 3.54 |
ENSMUST00000135267.1
ENSMUST00000052629.6 ENSMUST00000111235.2 |
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr10_+_63100156 | 3.50 |
ENSMUST00000044059.3
|
Atoh7
|
atonal homolog 7 (Drosophila) |
| chr16_-_15637277 | 3.48 |
ENSMUST00000023353.3
|
Mcm4
|
minichromosome maintenance deficient 4 homolog (S. cerevisiae) |
| chrX_-_51681703 | 3.36 |
ENSMUST00000088172.5
|
Hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr7_+_141475459 | 3.36 |
ENSMUST00000138092.1
ENSMUST00000146305.1 |
Tspan4
|
tetraspanin 4 |
| chrX_+_71555918 | 3.36 |
ENSMUST00000072699.6
ENSMUST00000114582.2 ENSMUST00000015361.4 ENSMUST00000088874.3 |
Hmgb3
|
high mobility group box 3 |
| chr17_-_71526819 | 3.35 |
ENSMUST00000024851.9
|
Ndc80
|
NDC80 homolog, kinetochore complex component (S. cerevisiae) |
| chr5_+_120649188 | 3.34 |
ENSMUST00000156722.1
|
Rasal1
|
RAS protein activator like 1 (GAP1 like) |
| chr14_+_31217850 | 3.33 |
ENSMUST00000090180.2
|
Sema3g
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G |
| chr16_-_23127702 | 3.30 |
ENSMUST00000115338.1
ENSMUST00000115337.1 ENSMUST00000023598.8 |
Rfc4
|
replication factor C (activator 1) 4 |
| chr11_-_101551837 | 3.30 |
ENSMUST00000017290.4
|
Brca1
|
breast cancer 1 |
| chr11_+_103649498 | 3.30 |
ENSMUST00000057870.2
|
Rprml
|
reprimo-like |
| chrX_-_51681856 | 3.27 |
ENSMUST00000114871.1
|
Hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr12_+_117843489 | 3.23 |
ENSMUST00000021592.9
|
Cdca7l
|
cell division cycle associated 7 like |
| chrX_-_60893430 | 3.22 |
ENSMUST00000135107.2
|
Sox3
|
SRY-box containing gene 3 |
| chr18_+_4921662 | 3.20 |
ENSMUST00000143254.1
|
Svil
|
supervillin |
| chr1_+_191063001 | 3.19 |
ENSMUST00000076952.5
ENSMUST00000139340.1 ENSMUST00000078259.6 |
Nsl1
|
NSL1, MIND kinetochore complex component, homolog (S. cerevisiae) |
| chr6_-_145076106 | 3.18 |
ENSMUST00000111742.1
ENSMUST00000048252.4 |
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr10_-_128704978 | 3.17 |
ENSMUST00000026416.7
ENSMUST00000026415.7 |
Cdk2
|
cyclin-dependent kinase 2 |
| chr8_+_105348163 | 3.16 |
ENSMUST00000073149.5
|
Slc9a5
|
solute carrier family 9 (sodium/hydrogen exchanger), member 5 |
| chr1_+_172481788 | 3.15 |
ENSMUST00000127052.1
|
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr7_-_48881596 | 3.14 |
ENSMUST00000119223.1
|
E2f8
|
E2F transcription factor 8 |
| chr17_-_29264115 | 3.11 |
ENSMUST00000024802.8
|
Ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr9_-_35116804 | 3.11 |
ENSMUST00000034537.6
|
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr5_+_33721724 | 3.10 |
ENSMUST00000067150.7
ENSMUST00000169212.2 ENSMUST00000114411.2 ENSMUST00000164207.3 |
Fgfr3
|
fibroblast growth factor receptor 3 |
| chr7_-_48881032 | 3.05 |
ENSMUST00000058745.8
|
E2f8
|
E2F transcription factor 8 |
| chr10_+_3366125 | 3.04 |
ENSMUST00000043374.5
|
Ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14c |
| chr5_-_138170992 | 3.01 |
ENSMUST00000139983.1
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr15_-_55090422 | 3.00 |
ENSMUST00000110231.1
ENSMUST00000023059.6 |
Dscc1
|
defective in sister chromatid cohesion 1 homolog (S. cerevisiae) |
| chr8_-_53638945 | 2.94 |
ENSMUST00000047768.4
|
Neil3
|
nei like 3 (E. coli) |
| chr14_-_67715585 | 2.93 |
ENSMUST00000163100.1
ENSMUST00000132705.1 ENSMUST00000124045.1 |
Cdca2
|
cell division cycle associated 2 |
| chr3_+_123446913 | 2.89 |
ENSMUST00000029603.8
|
Prss12
|
protease, serine, 12 neurotrypsin (motopsin) |
| chr18_+_56707725 | 2.88 |
ENSMUST00000025486.8
|
Lmnb1
|
lamin B1 |
| chr4_+_11558914 | 2.87 |
ENSMUST00000178703.1
ENSMUST00000095145.5 ENSMUST00000108306.2 ENSMUST00000070755.6 |
Rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr2_-_119618455 | 2.86 |
ENSMUST00000123818.1
|
Oip5
|
Opa interacting protein 5 |
| chr9_+_44084944 | 2.82 |
ENSMUST00000176416.1
ENSMUST00000065461.7 |
Usp2
|
ubiquitin specific peptidase 2 |
| chr11_-_86201144 | 2.82 |
ENSMUST00000044423.3
|
Brip1
|
BRCA1 interacting protein C-terminal helicase 1 |
| chrX_-_93632113 | 2.82 |
ENSMUST00000006856.2
|
Pola1
|
polymerase (DNA directed), alpha 1 |
| chrX_-_111463103 | 2.77 |
ENSMUST00000137712.2
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr5_-_138171216 | 2.76 |
ENSMUST00000147920.1
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr9_-_97018823 | 2.74 |
ENSMUST00000055433.4
|
Spsb4
|
splA/ryanodine receptor domain and SOCS box containing 4 |
| chr4_-_133968611 | 2.73 |
ENSMUST00000102552.1
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr6_+_117916981 | 2.73 |
ENSMUST00000179478.1
|
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chr15_-_55557748 | 2.72 |
ENSMUST00000172387.1
|
Mrpl13
|
mitochondrial ribosomal protein L13 |
| chr5_+_115845229 | 2.67 |
ENSMUST00000137952.1
ENSMUST00000148245.1 |
Cit
|
citron |
| chr2_-_30801698 | 2.65 |
ENSMUST00000050003.8
|
1700001O22Rik
|
RIKEN cDNA 1700001O22 gene |
| chr6_+_4755327 | 2.63 |
ENSMUST00000176551.1
|
Peg10
|
paternally expressed 10 |
| chr16_-_57606816 | 2.62 |
ENSMUST00000114371.3
|
Cmss1
|
cms small ribosomal subunit 1 |
| chr7_+_65862029 | 2.62 |
ENSMUST00000055576.5
ENSMUST00000098391.4 |
Pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr3_+_40800013 | 2.60 |
ENSMUST00000026858.5
ENSMUST00000170825.1 |
Plk4
|
polo-like kinase 4 |
| chr5_-_92435114 | 2.58 |
ENSMUST00000135112.1
|
Nup54
|
nucleoporin 54 |
| chr2_+_119618717 | 2.57 |
ENSMUST00000028771.7
|
Nusap1
|
nucleolar and spindle associated protein 1 |
| chr6_-_47594967 | 2.55 |
ENSMUST00000081721.6
ENSMUST00000114618.1 ENSMUST00000114616.1 |
Ezh2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr2_+_92599671 | 2.54 |
ENSMUST00000065797.6
|
Chst1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr19_-_10203880 | 2.54 |
ENSMUST00000142241.1
ENSMUST00000116542.2 ENSMUST00000025651.5 ENSMUST00000156291.1 |
Fen1
|
flap structure specific endonuclease 1 |
| chr10_+_128015157 | 2.54 |
ENSMUST00000178041.1
ENSMUST00000026461.7 |
Prim1
|
DNA primase, p49 subunit |
| chr12_-_11265768 | 2.51 |
ENSMUST00000166117.1
|
Gen1
|
Gen homolog 1, endonuclease (Drosophila) |
| chr6_+_117917281 | 2.50 |
ENSMUST00000180020.1
ENSMUST00000177570.1 |
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chr8_-_111300222 | 2.49 |
ENSMUST00000038739.4
|
Rfwd3
|
ring finger and WD repeat domain 3 |
| chr1_+_153425162 | 2.48 |
ENSMUST00000042373.5
|
Shcbp1l
|
Shc SH2-domain binding protein 1-like |
| chr2_-_66124994 | 2.47 |
ENSMUST00000028378.3
|
Galnt3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 |
| chr19_-_42431778 | 2.47 |
ENSMUST00000048630.6
|
Crtac1
|
cartilage acidic protein 1 |
| chr11_-_11808923 | 2.46 |
ENSMUST00000109664.1
ENSMUST00000150714.1 ENSMUST00000047689.4 ENSMUST00000171938.1 ENSMUST00000171080.1 |
Fignl1
|
fidgetin-like 1 |
| chr1_-_128359610 | 2.45 |
ENSMUST00000027601.4
|
Mcm6
|
minichromosome maintenance deficient 6 (MIS5 homolog, S. pombe) (S. cerevisiae) |
| chr2_+_72476159 | 2.44 |
ENSMUST00000102691.4
|
Cdca7
|
cell division cycle associated 7 |
| chrX_+_71556874 | 2.43 |
ENSMUST00000123100.1
|
Hmgb3
|
high mobility group box 3 |
| chr10_-_7212222 | 2.42 |
ENSMUST00000015346.5
|
Cnksr3
|
Cnksr family member 3 |
| chr12_+_24708984 | 2.42 |
ENSMUST00000154588.1
|
Rrm2
|
ribonucleotide reductase M2 |
| chr4_-_138396438 | 2.41 |
ENSMUST00000105032.2
|
Fam43b
|
family with sequence similarity 43, member B |
| chr5_-_138171813 | 2.40 |
ENSMUST00000155902.1
ENSMUST00000148879.1 |
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr15_-_98004634 | 2.39 |
ENSMUST00000131560.1
ENSMUST00000088355.5 |
Col2a1
|
collagen, type II, alpha 1 |
| chr1_-_21961942 | 2.39 |
ENSMUST00000115300.1
|
Kcnq5
|
potassium voltage-gated channel, subfamily Q, member 5 |
| chr4_+_108579445 | 2.38 |
ENSMUST00000102744.3
|
Orc1
|
origin recognition complex, subunit 1 |
| chr4_-_116123618 | 2.37 |
ENSMUST00000102704.3
ENSMUST00000102705.3 |
Rad54l
|
RAD54 like (S. cerevisiae) |
| chr2_+_158768083 | 2.32 |
ENSMUST00000029183.2
|
Fam83d
|
family with sequence similarity 83, member D |
| chr11_-_82991137 | 2.31 |
ENSMUST00000138797.1
|
Slfn9
|
schlafen 9 |
| chrX_-_52613913 | 2.30 |
ENSMUST00000069360.7
|
Gpc3
|
glypican 3 |
| chrY_+_90785442 | 2.30 |
ENSMUST00000177591.1
ENSMUST00000177671.1 ENSMUST00000179077.1 |
Erdr1
|
erythroid differentiation regulator 1 |
| chr1_-_33669745 | 2.30 |
ENSMUST00000027312.9
|
Prim2
|
DNA primase, p58 subunit |
| chr17_-_35838208 | 2.30 |
ENSMUST00000134978.2
|
Tubb5
|
tubulin, beta 5 class I |
| chr15_+_55557575 | 2.30 |
ENSMUST00000170046.1
|
Mtbp
|
Mdm2, transformed 3T3 cell double minute p53 binding protein |
| chr5_+_106964319 | 2.30 |
ENSMUST00000031221.5
ENSMUST00000117196.2 ENSMUST00000076467.6 |
Cdc7
|
cell division cycle 7 (S. cerevisiae) |
| chr13_-_92794809 | 2.28 |
ENSMUST00000022213.7
|
Thbs4
|
thrombospondin 4 |
| chr3_+_40800054 | 2.28 |
ENSMUST00000168287.1
|
Plk4
|
polo-like kinase 4 |
| chr13_+_73467197 | 2.27 |
ENSMUST00000022099.8
|
Lpcat1
|
lysophosphatidylcholine acyltransferase 1 |
| chr7_+_141475240 | 2.27 |
ENSMUST00000026585.7
|
Tspan4
|
tetraspanin 4 |
| chr17_+_56040350 | 2.24 |
ENSMUST00000002914.8
|
Chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr11_+_100334407 | 2.22 |
ENSMUST00000017309.1
|
Gast
|
gastrin |
| chr11_-_34833631 | 2.19 |
ENSMUST00000093191.2
|
Spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr17_+_56304313 | 2.18 |
ENSMUST00000113035.1
ENSMUST00000113039.2 ENSMUST00000142387.1 |
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr11_-_97629685 | 2.17 |
ENSMUST00000052281.4
|
E130012A19Rik
|
RIKEN cDNA E130012A19 gene |
| chr11_+_80089385 | 2.17 |
ENSMUST00000108239.1
ENSMUST00000017694.5 |
Atad5
|
ATPase family, AAA domain containing 5 |
| chr5_-_138172383 | 2.16 |
ENSMUST00000000505.9
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr19_+_38930909 | 2.16 |
ENSMUST00000025965.5
|
Hells
|
helicase, lymphoid specific |
| chr7_-_44548733 | 2.12 |
ENSMUST00000145956.1
ENSMUST00000049343.8 |
Pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr2_+_109280738 | 2.11 |
ENSMUST00000028527.7
|
Kif18a
|
kinesin family member 18A |
| chr16_+_17070220 | 2.11 |
ENSMUST00000141959.1
|
Ypel1
|
yippee-like 1 (Drosophila) |
| chr5_-_151651216 | 2.11 |
ENSMUST00000038131.9
|
Rfc3
|
replication factor C (activator 1) 3 |
| chr8_-_78508876 | 2.11 |
ENSMUST00000049245.7
|
Rbmxl1
|
RNA binding motif protein, X linked-like-1 |
| chr19_+_38931008 | 2.08 |
ENSMUST00000145051.1
|
Hells
|
helicase, lymphoid specific |
| chr9_-_123260776 | 2.07 |
ENSMUST00000068140.4
|
Tmem158
|
transmembrane protein 158 |
| chr9_+_51765325 | 2.06 |
ENSMUST00000065496.5
|
Arhgap20
|
Rho GTPase activating protein 20 |
| chr17_-_40935047 | 2.04 |
ENSMUST00000087114.3
|
Cenpq
|
centromere protein Q |
| chr9_-_106656081 | 2.03 |
ENSMUST00000023959.7
|
Grm2
|
glutamate receptor, metabotropic 2 |
| chr16_+_17070281 | 2.03 |
ENSMUST00000090199.3
|
Ypel1
|
yippee-like 1 (Drosophila) |
| chr10_-_127666598 | 2.01 |
ENSMUST00000099157.3
|
Nab2
|
Ngfi-A binding protein 2 |
| chr8_-_124434323 | 2.01 |
ENSMUST00000140012.1
|
Pgbd5
|
piggyBac transposable element derived 5 |
| chr17_+_35861343 | 2.01 |
ENSMUST00000172931.1
|
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr2_-_125625065 | 2.00 |
ENSMUST00000089776.2
|
Cep152
|
centrosomal protein 152 |
| chr15_-_98004695 | 1.99 |
ENSMUST00000023123.8
|
Col2a1
|
collagen, type II, alpha 1 |
| chrX_+_134059315 | 1.98 |
ENSMUST00000144483.1
|
Cstf2
|
cleavage stimulation factor, 3' pre-RNA subunit 2 |
| chr15_-_64382908 | 1.97 |
ENSMUST00000177374.1
ENSMUST00000023008.9 ENSMUST00000110115.2 ENSMUST00000110114.3 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr2_+_151702182 | 1.96 |
ENSMUST00000109872.1
|
Tmem74b
|
transmembrane protein 74b |
| chr17_+_35841491 | 1.95 |
ENSMUST00000082337.6
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr2_+_72476225 | 1.95 |
ENSMUST00000157019.1
|
Cdca7
|
cell division cycle associated 7 |
| chr15_-_64382736 | 1.94 |
ENSMUST00000176384.1
ENSMUST00000175799.1 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr19_+_46075842 | 1.94 |
ENSMUST00000165017.1
|
Nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr17_-_35838259 | 1.93 |
ENSMUST00000001566.8
|
Tubb5
|
tubulin, beta 5 class I |
| chr11_+_69015911 | 1.91 |
ENSMUST00000021278.7
ENSMUST00000161455.1 ENSMUST00000116359.2 |
Ctc1
|
CTS telomere maintenance complex component 1 |
| chr5_-_8422582 | 1.91 |
ENSMUST00000168500.1
ENSMUST00000002368.9 |
Dbf4
|
DBF4 homolog (S. cerevisiae) |
| chr17_-_25115905 | 1.90 |
ENSMUST00000024987.5
ENSMUST00000115181.2 |
Telo2
|
TEL2, telomere maintenance 2, homolog (S. cerevisiae) |
| chr18_+_36760214 | 1.89 |
ENSMUST00000049323.7
|
Wdr55
|
WD repeat domain 55 |
| chr1_+_172312367 | 1.88 |
ENSMUST00000039506.9
|
Igsf8
|
immunoglobulin superfamily, member 8 |
| chr10_-_81001338 | 1.87 |
ENSMUST00000099462.1
ENSMUST00000118233.1 |
Gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr5_-_8422695 | 1.86 |
ENSMUST00000171808.1
|
Dbf4
|
DBF4 homolog (S. cerevisiae) |
| chr6_-_8259098 | 1.85 |
ENSMUST00000012627.4
|
Rpa3
|
replication protein A3 |
| chr5_+_112343068 | 1.85 |
ENSMUST00000112359.2
ENSMUST00000035279.3 |
Hps4
|
Hermansky-Pudlak syndrome 4 homolog (human) |
| chrX_-_52613936 | 1.84 |
ENSMUST00000114857.1
|
Gpc3
|
glypican 3 |
| chr2_-_34913976 | 1.82 |
ENSMUST00000028232.3
|
Phf19
|
PHD finger protein 19 |
| chr11_-_116335384 | 1.80 |
ENSMUST00000036215.7
|
Foxj1
|
forkhead box J1 |
| chr8_-_105851981 | 1.79 |
ENSMUST00000040776.4
|
Cenpt
|
centromere protein T |
| chr7_-_142578093 | 1.78 |
ENSMUST00000149974.1
ENSMUST00000152754.1 |
H19
|
H19 fetal liver mRNA |
| chr11_-_6606053 | 1.78 |
ENSMUST00000045713.3
|
Nacad
|
NAC alpha domain containing |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.5 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 2.0 | 10.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 2.0 | 20.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 1.9 | 9.7 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 1.9 | 7.7 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 1.6 | 4.7 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 1.5 | 4.6 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 1.5 | 4.5 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 1.5 | 4.5 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 1.4 | 5.7 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 1.4 | 2.8 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 1.4 | 4.2 | GO:0071163 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 1.3 | 20.1 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 1.3 | 3.8 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 1.2 | 11.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 1.2 | 13.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 1.2 | 5.8 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 1.1 | 7.9 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 1.1 | 4.5 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 1.1 | 6.6 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 1.1 | 7.7 | GO:0044838 | cell quiescence(GO:0044838) |
| 1.1 | 26.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 1.1 | 3.3 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 1.1 | 5.4 | GO:0072138 | mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 1.0 | 6.9 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 1.0 | 2.9 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.9 | 2.8 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.9 | 3.6 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.9 | 2.6 | GO:1901254 | regulation of translation at synapse, modulating synaptic transmission(GO:0099547) regulation of translation at postsynapse, modulating synaptic transmission(GO:0099578) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.9 | 5.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.8 | 0.8 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.8 | 15.0 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.8 | 4.0 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.8 | 9.2 | GO:0033504 | floor plate development(GO:0033504) |
| 0.8 | 11.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.8 | 2.3 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.8 | 3.8 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.7 | 1.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.7 | 2.1 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.7 | 6.2 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.7 | 2.7 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.6 | 1.9 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.6 | 8.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.6 | 3.7 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.6 | 1.2 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.6 | 2.9 | GO:0032902 | nerve growth factor production(GO:0032902) |
| 0.6 | 1.7 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.6 | 8.5 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.6 | 1.7 | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation(GO:0051316) |
| 0.5 | 3.2 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.5 | 1.5 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.5 | 1.5 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.5 | 2.9 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.5 | 2.4 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.5 | 1.4 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.5 | 1.9 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.5 | 1.4 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.5 | 7.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.5 | 4.6 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.5 | 0.5 | GO:0032240 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.5 | 5.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.4 | 0.9 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.4 | 4.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.4 | 0.9 | GO:0051311 | meiotic metaphase plate congression(GO:0051311) |
| 0.4 | 1.8 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.4 | 6.5 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.4 | 1.3 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.4 | 4.3 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.4 | 0.9 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.4 | 1.3 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.4 | 2.5 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.4 | 6.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.4 | 0.4 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.4 | 2.9 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.4 | 1.2 | GO:1904020 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.4 | 3.2 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.4 | 0.4 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.4 | 4.4 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.4 | 1.6 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.4 | 2.8 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.4 | 1.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.4 | 4.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.4 | 0.4 | GO:0090172 | microtubule cytoskeleton organization involved in homologous chromosome segregation(GO:0090172) |
| 0.4 | 1.1 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.4 | 3.7 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.4 | 1.1 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 0.4 | 1.8 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.4 | 1.8 | GO:0061141 | lung ciliated cell differentiation(GO:0061141) |
| 0.4 | 3.2 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.3 | 0.3 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.3 | 1.0 | GO:0061110 | dense core granule biogenesis(GO:0061110) |
| 0.3 | 1.0 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.3 | 1.0 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.3 | 4.4 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.3 | 7.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.3 | 0.7 | GO:0061086 | negative regulation of histone phosphorylation(GO:0033128) negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.3 | 2.3 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.3 | 1.9 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.3 | 1.0 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.3 | 1.3 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.3 | 1.2 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.3 | 1.5 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.3 | 0.6 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.3 | 1.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 0.9 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.3 | 2.1 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.3 | 3.6 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.3 | 0.9 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.3 | 2.3 | GO:1904667 | negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.3 | 0.6 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.3 | 2.9 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.3 | 1.2 | GO:1990401 | embryonic lung development(GO:1990401) |
| 0.3 | 2.0 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.3 | 1.1 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.3 | 1.4 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.3 | 0.6 | GO:0072708 | response to sorbitol(GO:0072708) |
| 0.3 | 1.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.3 | 0.5 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.3 | 1.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.3 | 1.6 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.3 | 1.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.3 | 0.8 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.3 | 5.5 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.3 | 0.8 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.3 | 0.5 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.3 | 2.1 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.3 | 0.8 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.3 | 3.0 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.3 | 2.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.3 | 1.3 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.2 | 0.7 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.2 | 0.7 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.2 | 0.5 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.2 | 1.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.2 | 0.5 | GO:0019042 | viral latency(GO:0019042) establishment of viral latency(GO:0019043) |
| 0.2 | 0.5 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.2 | 1.6 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.2 | 2.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.2 | 1.8 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 0.7 | GO:1904690 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.2 | 0.7 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.2 | 0.9 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 | 0.7 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.2 | 2.1 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.2 | 0.8 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.2 | 0.6 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.2 | 1.0 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.2 | 0.4 | GO:0048296 | regulation of isotype switching to IgA isotypes(GO:0048296) positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.2 | 0.2 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.2 | 2.5 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.2 | 1.0 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.2 | 8.3 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.2 | 3.0 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.2 | 0.8 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.2 | 0.8 | GO:1904799 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) |
| 0.2 | 2.3 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.2 | 1.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.2 | 3.7 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.2 | 4.0 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 4.2 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.2 | 9.1 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.2 | 0.2 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.2 | 0.2 | GO:0072368 | regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 0.2 | 1.5 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.2 | 1.5 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.2 | 0.7 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.2 | 0.9 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.2 | 0.7 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 0.7 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.2 | 1.4 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.2 | 5.0 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.2 | 1.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 0.5 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.2 | 0.5 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.2 | 13.1 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.2 | 7.9 | GO:0033045 | regulation of sister chromatid segregation(GO:0033045) |
| 0.2 | 0.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 0.5 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.2 | 3.3 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.2 | 0.7 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.2 | 1.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.2 | 0.8 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.2 | 2.0 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 0.5 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.2 | 0.3 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.2 | 3.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.2 | 4.2 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.1 | 0.7 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 0.1 | 4.0 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 1.0 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 1.5 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 1.7 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 3.5 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 0.6 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.1 | 3.6 | GO:1903859 | regulation of dendrite extension(GO:1903859) |
| 0.1 | 0.3 | GO:0048627 | myoblast development(GO:0048627) |
| 0.1 | 5.9 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.1 | 1.1 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.1 | 0.8 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.1 | 1.9 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 1.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.9 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 | 2.3 | GO:1904814 | regulation of protein localization to chromosome, telomeric region(GO:1904814) positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.1 | 2.2 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 0.9 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.4 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.1 | 0.2 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.1 | 0.2 | GO:0072683 | T cell extravasation(GO:0072683) regulation of T cell extravasation(GO:2000407) positive regulation of T cell extravasation(GO:2000409) |
| 0.1 | 0.7 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 1.2 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 0.6 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 0.5 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 0.6 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 0.6 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 1.7 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 1.4 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.1 | 1.0 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.1 | 0.9 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 2.2 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 1.3 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.1 | 0.3 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 | 0.3 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.1 | 0.4 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.1 | 0.7 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.8 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.3 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.1 | 2.4 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 0.5 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.1 | 0.7 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.1 | 0.7 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.1 | 1.0 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 0.3 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 | 0.6 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.4 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 0.5 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 | 0.7 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.3 | GO:1903351 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.1 | 2.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.1 | 0.4 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.3 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.1 | 0.5 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 1.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.7 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 1.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 1.6 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 1.6 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.8 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 3.7 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 1.7 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.1 | 0.4 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 0.2 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.4 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.1 | 0.8 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.1 | 1.2 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 1.3 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.5 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.3 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 0.4 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.8 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.3 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 3.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.4 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.1 | 1.1 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 1.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 2.0 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.8 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 2.2 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 5.4 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 3.3 | GO:0030890 | positive regulation of B cell proliferation(GO:0030890) |
| 0.1 | 0.4 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 1.0 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.1 | 0.8 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.1 | 2.9 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.1 | 0.5 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 | 0.4 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.1 | 1.5 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.1 | 0.2 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.4 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.1 | 0.2 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 | 1.3 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.7 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 0.4 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 1.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.1 | GO:0061209 | cell proliferation involved in mesonephros development(GO:0061209) |
| 0.1 | 1.0 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.3 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.1 | 0.8 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.9 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 0.5 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.2 | GO:0060447 | peripheral nervous system axon regeneration(GO:0014012) bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 | 3.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.3 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) epithelial cell proliferation involved in prostate gland development(GO:0060767) regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 0.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.1 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.1 | 1.0 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.1 | 0.5 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.5 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 0.2 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.1 | 0.5 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 0.2 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.1 | 0.2 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.1 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.2 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.1 | 0.2 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 0.2 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.1 | 0.3 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.1 | 0.2 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 5.6 | GO:0071897 | DNA biosynthetic process(GO:0071897) |
| 0.1 | 0.8 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.1 | 0.3 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 0.2 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) histone H3-S10 phosphorylation(GO:0043987) |
| 0.1 | 1.1 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.1 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 0.5 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.1 | 0.2 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.1 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.7 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.1 | 0.6 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 | 0.5 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 1.1 | GO:0051654 | establishment of mitochondrion localization(GO:0051654) |
| 0.1 | 8.2 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.1 | 0.1 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 2.4 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 1.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 0.3 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 | 0.5 | GO:0006569 | tryptophan metabolic process(GO:0006568) tryptophan catabolic process(GO:0006569) indolalkylamine metabolic process(GO:0006586) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) kynurenine metabolic process(GO:0070189) |
| 0.1 | 0.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.5 | GO:0032682 | negative regulation of chemokine production(GO:0032682) |
| 0.0 | 0.3 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.0 | GO:0072710 | response to hydroxyurea(GO:0072710) |
| 0.0 | 0.6 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.3 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.0 | 0.4 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 1.5 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.1 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.3 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.4 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.5 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.2 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0003360 | brainstem development(GO:0003360) |
| 0.0 | 0.5 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.9 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 4.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.0 | 0.4 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.6 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.1 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.0 | 1.7 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.5 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.0 | 0.1 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 1.3 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.1 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.3 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.2 | GO:0034398 | telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 2.8 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.5 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.5 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.3 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 1.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.2 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) positive regulation of glycogen catabolic process(GO:0045819) |
| 0.0 | 0.3 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.0 | 0.4 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.3 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.8 | GO:0052646 | alditol phosphate metabolic process(GO:0052646) |
| 0.0 | 0.9 | GO:1903831 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 0.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.1 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.0 | 0.5 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.3 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.1 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.4 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 1.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.9 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.4 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.2 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 1.2 | GO:0046427 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.0 | 1.5 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.4 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.1 | GO:0002194 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.0 | 0.5 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 1.4 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.7 | GO:0071398 | cellular response to fatty acid(GO:0071398) |
| 0.0 | 0.1 | GO:1903936 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.5 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.2 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.2 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.4 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.2 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.2 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.5 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.1 | GO:1905169 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.7 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.4 | GO:0045624 | positive regulation of T-helper cell differentiation(GO:0045624) |
| 0.0 | 0.4 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.3 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 2.0 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.5 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 0.1 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.0 | 0.1 | GO:0009146 | purine nucleoside triphosphate catabolic process(GO:0009146) |
| 0.0 | 0.6 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.2 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.9 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.3 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.6 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.6 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.8 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.3 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.3 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.6 | GO:0007492 | endoderm development(GO:0007492) |
| 0.0 | 0.3 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.5 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.1 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 1.1 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 0.7 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.1 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.1 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 | 0.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 1.7 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.4 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.1 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.2 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 12.8 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 2.5 | 7.6 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 2.5 | 37.6 | GO:0042555 | MCM complex(GO:0042555) |
| 2.0 | 6.0 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 1.8 | 17.9 | GO:0000796 | condensin complex(GO:0000796) |
| 1.6 | 11.0 | GO:0001652 | granular component(GO:0001652) |
| 1.5 | 7.7 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 1.4 | 5.7 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.4 | 4.2 | GO:1990423 | RZZ complex(GO:1990423) |
| 1.4 | 4.2 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 1.3 | 7.7 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 1.3 | 7.6 | GO:0098536 | deuterosome(GO:0098536) |
| 1.0 | 5.1 | GO:0031523 | Myb complex(GO:0031523) |
| 1.0 | 2.9 | GO:0071920 | cleavage body(GO:0071920) |
| 0.9 | 4.5 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.8 | 3.3 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.8 | 3.3 | GO:0090537 | CERF complex(GO:0090537) |
| 0.8 | 4.8 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.8 | 4.7 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.7 | 3.7 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.7 | 3.7 | GO:0071547 | piP-body(GO:0071547) |
| 0.7 | 2.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.7 | 3.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.6 | 2.6 | GO:0019034 | viral replication complex(GO:0019034) dendritic filopodium(GO:1902737) |
| 0.6 | 5.4 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.6 | 0.6 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.6 | 5.8 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.6 | 4.0 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.6 | 4.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.5 | 3.3 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.5 | 3.8 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.5 | 4.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.5 | 4.2 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.5 | 1.5 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.5 | 3.6 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.5 | 3.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.5 | 1.8 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.4 | 4.4 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.4 | 2.6 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.4 | 2.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.4 | 7.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.4 | 3.7 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.4 | 2.0 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.4 | 1.6 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.4 | 1.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.4 | 7.9 | GO:0010369 | chromocenter(GO:0010369) |
| 0.4 | 1.1 | GO:0031533 | mRNA cap methyltransferase complex(GO:0031533) |
| 0.4 | 5.7 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.4 | 4.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.4 | 2.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.3 | 0.3 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.3 | 1.3 | GO:1990707 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.3 | 19.5 | GO:0005657 | replication fork(GO:0005657) |
| 0.3 | 3.7 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.3 | 1.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.3 | 7.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 1.7 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.3 | 4.8 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.3 | 0.8 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.3 | 1.4 | GO:0001740 | Barr body(GO:0001740) |
| 0.3 | 1.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.3 | 1.6 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.3 | 4.0 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.2 | 3.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 1.7 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.2 | 2.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.2 | 2.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.2 | 3.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 1.6 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.2 | 1.6 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.2 | 0.7 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.2 | 2.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.2 | 0.8 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 0.8 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.2 | 0.6 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.2 | 0.8 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.2 | 0.6 | GO:0031417 | NatC complex(GO:0031417) |
| 0.2 | 0.9 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 0.7 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.2 | 0.6 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.2 | 3.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 1.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 0.7 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.2 | 1.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.2 | 1.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.2 | 1.8 | GO:0032797 | SMN complex(GO:0032797) |
| 0.2 | 9.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 1.9 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.2 | 6.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 0.6 | GO:0008623 | CHRAC(GO:0008623) |
| 0.2 | 1.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.2 | 1.0 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.2 | 1.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 23.9 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 3.7 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.5 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 0.3 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 1.2 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.1 | 0.5 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.1 | 2.5 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 0.5 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.4 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.1 | 1.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.6 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 3.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.5 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 1.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 11.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 2.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.8 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 10.9 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.5 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 0.6 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 16.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 5.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.7 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 4.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 8.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 0.4 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 1.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 1.0 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 0.3 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.7 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 2.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 0.8 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.7 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 2.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 4.2 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 1.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.5 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 0.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.2 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 0.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 1.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 2.0 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 0.7 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 5.4 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 2.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.2 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.1 | 0.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.2 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.3 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 0.4 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 2.3 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.4 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.4 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 6.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.7 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 1.0 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 1.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 3.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 1.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 4.0 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.4 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 8.6 | GO:0043292 | contractile fiber(GO:0043292) |
| 0.0 | 0.1 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 3.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 1.4 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.8 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 1.3 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.9 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 3.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 2.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.3 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 2.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 1.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0070877 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 3.4 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 7.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.5 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.1 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.2 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 1.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 16.4 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 2.6 | 12.8 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 2.4 | 9.7 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 2.3 | 13.9 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 1.7 | 5.1 | GO:0032142 | single guanine insertion binding(GO:0032142) |
| 1.7 | 6.6 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 1.5 | 4.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 1.5 | 7.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 1.4 | 4.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 1.3 | 2.6 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 1.1 | 3.3 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 1.1 | 3.3 | GO:0070615 | nucleosome-dependent ATPase activity(GO:0070615) |
| 1.0 | 3.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 1.0 | 11.0 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.9 | 3.6 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.9 | 3.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.8 | 4.0 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.8 | 4.0 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.8 | 3.1 | GO:0047288 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.8 | 3.8 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.7 | 2.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.6 | 8.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.6 | 2.5 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.6 | 4.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.6 | 23.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.6 | 3.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.6 | 4.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.6 | 5.5 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.6 | 4.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.6 | 4.7 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.6 | 1.7 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.5 | 3.2 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.5 | 3.7 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.5 | 4.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.5 | 2.5 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.5 | 2.0 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.5 | 2.4 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.4 | 3.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.4 | 2.6 | GO:0097617 | annealing activity(GO:0097617) |
| 0.4 | 4.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.4 | 1.3 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.4 | 2.5 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.4 | 1.1 | GO:0004482 | mRNA (guanine-N7-)-methyltransferase activity(GO:0004482) |
| 0.4 | 1.1 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.4 | 1.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.4 | 1.8 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.3 | 9.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.3 | 1.0 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.3 | 2.4 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.3 | 3.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 1.6 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.3 | 1.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.3 | 1.3 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.3 | 1.3 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.3 | 2.8 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.3 | 1.2 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.3 | 1.2 | GO:0004058 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 0.3 | 0.9 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.3 | 1.8 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.3 | 0.9 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.3 | 1.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 0.3 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.3 | 2.3 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.3 | 1.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.3 | 2.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.3 | 4.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 1.0 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) |
| 0.2 | 1.5 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.2 | 3.4 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.2 | 1.6 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.2 | 0.7 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 6.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 3.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 7.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.2 | 0.7 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.2 | 1.8 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.2 | 0.9 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.2 | 1.3 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.2 | 1.7 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.2 | 1.7 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 4.9 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.2 | 3.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 1.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 1.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.2 | 1.8 | GO:0098505 | single-stranded telomeric DNA binding(GO:0043047) G-rich strand telomeric DNA binding(GO:0098505) |
| 0.2 | 0.8 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.2 | 4.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 1.7 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 1.5 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 1.7 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.2 | 0.6 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.2 | 1.4 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.2 | 3.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 8.0 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.2 | 5.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 1.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.2 | 2.6 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.2 | 1.7 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.2 | 4.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 3.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 1.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 1.4 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 3.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 1.0 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.2 | 1.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 3.0 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.2 | 1.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 1.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 0.9 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.2 | 0.9 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 1.3 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 1.7 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.9 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.9 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 4.7 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 0.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 10.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 0.5 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 3.6 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.3 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 3.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.5 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 1.8 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 16.9 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 1.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 1.8 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.5 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.1 | 0.9 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.1 | 0.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.4 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 6.4 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 0.3 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 0.5 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.6 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 0.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.7 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.5 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.1 | 1.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.5 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 4.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 1.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 4.0 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 22.6 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 1.1 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.1 | 0.7 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 3.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 0.3 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.1 | 3.7 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 1.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 2.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.3 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) TFIIIC-class transcription factor binding(GO:0001156) |
| 0.1 | 0.4 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 1.5 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 1.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 0.7 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.4 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 0.6 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.4 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.1 | 0.8 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.1 | 5.6 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 0.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 12.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.9 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.3 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.1 | 0.4 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 2.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 2.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 0.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 5.3 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 6.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.2 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.1 | 0.5 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 1.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.3 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.1 | 0.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 0.1 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 1.0 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.1 | 0.5 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.1 | 2.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 1.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 4.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 10.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 1.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.4 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 2.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.2 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 1.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.2 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 1.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.4 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 12.7 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.5 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 1.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.2 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.9 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.2 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 1.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 2.4 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.2 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 53.2 | PID ATR PATHWAY | ATR signaling pathway |
| 1.0 | 13.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.5 | 24.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.5 | 40.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.4 | 4.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.3 | 2.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.3 | 9.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.3 | 7.1 | PID ATM PATHWAY | ATM pathway |
| 0.3 | 1.0 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 6.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 3.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 2.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 7.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 5.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 12.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 5.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 2.0 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 4.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 2.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 1.3 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 1.5 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 2.0 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 2.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 0.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 1.9 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 3.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 13.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 1.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 2.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 2.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 2.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.9 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 1.3 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.8 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 35.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 1.4 | 26.6 | REACTOME LAGGING STRAND SYNTHESIS | Genes involved in Lagging Strand Synthesis |
| 1.1 | 17.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 1.0 | 17.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 1.0 | 13.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 1.0 | 1.0 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.5 | 6.3 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.5 | 4.8 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.5 | 10.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.5 | 5.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.4 | 1.8 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.4 | 9.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.3 | 8.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.3 | 6.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.3 | 4.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 5.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.3 | 0.5 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.2 | 1.7 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.2 | 9.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.2 | 2.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 7.8 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.2 | 1.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 11.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 5.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 2.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 9.0 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 2.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 16.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 2.7 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.2 | 3.0 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 3.0 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.2 | 3.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 0.2 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.2 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 0.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 5.1 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 2.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 9.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.0 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.5 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 2.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 0.8 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 1.9 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 1.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.0 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 2.6 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 5.5 | REACTOME MITOTIC G2 G2 M PHASES | Genes involved in Mitotic G2-G2/M phases |
| 0.1 | 1.6 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.8 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 3.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.5 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 2.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.5 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 2.6 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 3.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 4.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.6 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.1 | 0.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 1.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 4.9 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.0 | 9.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 1.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 2.8 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.6 | REACTOME TRANSCRIPTION | Genes involved in Transcription |
| 0.0 | 0.9 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 2.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 2.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.5 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 0.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |