Project
avrg: 12D miR HR13_24
Navigation
Downloads
Results for Hcfc1_Six5_Smarcc2_Zfp143
Z-value: 7.35
Transcription factors associated with Hcfc1_Six5_Smarcc2_Zfp143
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
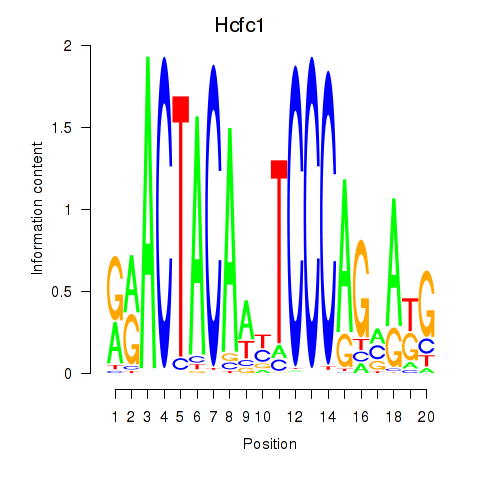
Hcfc1
|
ENSMUSG00000031386.8 | host cell factor C1 |
|
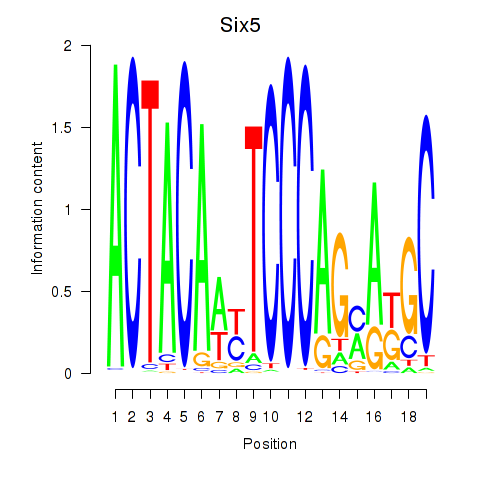
Six5
|
ENSMUSG00000040841.5 | sine oculis-related homeobox 5 |
|
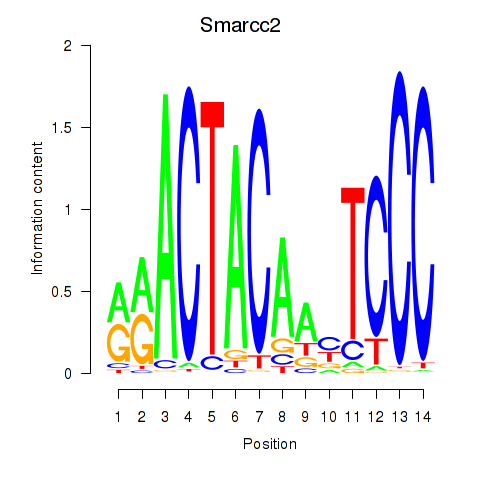
Smarcc2
|
ENSMUSG00000025369.8 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
|
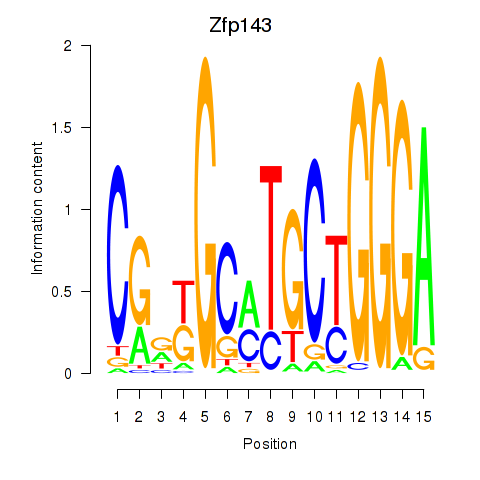
Zfp143
|
ENSMUSG00000061079.7 | zinc finger protein 143 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hcfc1 | mm10_v2_chrX_-_73966329_73966376 | 0.97 | 1.0e-06 | Click! |
| Zfp143 | mm10_v2_chr7_+_110061702_110061732 | 0.94 | 2.2e-05 | Click! |
| Smarcc2 | mm10_v2_chr10_+_128459236_128459248 | 0.91 | 9.4e-05 | Click! |
| Six5 | mm10_v2_chr7_+_19094594_19094633 | -0.69 | 1.8e-02 | Click! |
Activity profile of Hcfc1_Six5_Smarcc2_Zfp143 motif
Sorted Z-values of Hcfc1_Six5_Smarcc2_Zfp143 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrY_+_90784738 | 11.15 |
ENSMUST00000179483.1
|
Erdr1
|
erythroid differentiation regulator 1 |
| chr6_+_124829540 | 10.25 |
ENSMUST00000150120.1
|
Cdca3
|
cell division cycle associated 3 |
| chr2_+_118598209 | 9.50 |
ENSMUST00000038341.7
|
Bub1b
|
budding uninhibited by benzimidazoles 1 homolog, beta (S. cerevisiae) |
| chr2_+_152847961 | 8.14 |
ENSMUST00000164120.1
ENSMUST00000178997.1 ENSMUST00000109816.1 |
Tpx2
|
TPX2, microtubule-associated protein homolog (Xenopus laevis) |
| chr5_+_33658123 | 7.69 |
ENSMUST00000074849.6
ENSMUST00000079534.4 |
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr2_+_152847993 | 7.37 |
ENSMUST00000028969.8
|
Tpx2
|
TPX2, microtubule-associated protein homolog (Xenopus laevis) |
| chr7_+_29307924 | 7.31 |
ENSMUST00000108230.1
ENSMUST00000065181.5 |
Dpf1
|
D4, zinc and double PHD fingers family 1 |
| chr11_+_117849223 | 6.96 |
ENSMUST00000081387.4
|
Birc5
|
baculoviral IAP repeat-containing 5 |
| chr8_+_71464910 | 6.91 |
ENSMUST00000048914.6
|
Mrpl34
|
mitochondrial ribosomal protein L34 |
| chr6_+_124829582 | 6.69 |
ENSMUST00000024270.7
|
Cdca3
|
cell division cycle associated 3 |
| chr9_+_107587711 | 6.23 |
ENSMUST00000010192.5
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr7_+_29303958 | 6.15 |
ENSMUST00000049977.6
|
Dpf1
|
D4, zinc and double PHD fingers family 1 |
| chr11_+_117849286 | 6.14 |
ENSMUST00000093906.4
|
Birc5
|
baculoviral IAP repeat-containing 5 |
| chr11_-_101551837 | 6.09 |
ENSMUST00000017290.4
|
Brca1
|
breast cancer 1 |
| chr7_+_29303938 | 5.94 |
ENSMUST00000108231.1
|
Dpf1
|
D4, zinc and double PHD fingers family 1 |
| chr19_-_10203880 | 5.93 |
ENSMUST00000142241.1
ENSMUST00000116542.2 ENSMUST00000025651.5 ENSMUST00000156291.1 |
Fen1
|
flap structure specific endonuclease 1 |
| chr19_+_5024006 | 5.83 |
ENSMUST00000025826.5
|
Slc29a2
|
solute carrier family 29 (nucleoside transporters), member 2 |
| chr13_-_54590047 | 5.78 |
ENSMUST00000148222.1
ENSMUST00000026987.5 |
Nop16
|
NOP16 nucleolar protein |
| chr4_+_156109971 | 5.69 |
ENSMUST00000072554.6
ENSMUST00000169550.1 ENSMUST00000105576.1 |
9430015G10Rik
|
RIKEN cDNA 9430015G10 gene |
| chrY_+_90785442 | 5.53 |
ENSMUST00000177591.1
ENSMUST00000177671.1 ENSMUST00000179077.1 |
Erdr1
|
erythroid differentiation regulator 1 |
| chr4_-_117182623 | 5.46 |
ENSMUST00000065896.2
|
Kif2c
|
kinesin family member 2C |
| chr13_+_41249841 | 5.42 |
ENSMUST00000165561.2
|
Smim13
|
small integral membrane protein 13 |
| chr19_-_9899450 | 5.24 |
ENSMUST00000025562.7
|
Incenp
|
inner centromere protein |
| chr15_+_85859689 | 5.06 |
ENSMUST00000170629.1
|
Gtse1
|
G two S phase expressed protein 1 |
| chr12_-_91384403 | 4.95 |
ENSMUST00000141429.1
|
Cep128
|
centrosomal protein 128 |
| chr15_+_78428564 | 4.88 |
ENSMUST00000166142.2
ENSMUST00000162517.1 ENSMUST00000089414.4 |
Kctd17
|
potassium channel tetramerisation domain containing 17 |
| chr4_-_124936852 | 4.87 |
ENSMUST00000030690.5
ENSMUST00000084296.3 |
Cdca8
|
cell division cycle associated 8 |
| chr4_-_41275091 | 4.78 |
ENSMUST00000030143.6
ENSMUST00000108068.1 |
Ubap2
|
ubiquitin-associated protein 2 |
| chr15_+_102296256 | 4.67 |
ENSMUST00000064924.4
|
Espl1
|
extra spindle poles-like 1 (S. cerevisiae) |
| chr2_+_181680284 | 4.65 |
ENSMUST00000103042.3
|
Tcea2
|
transcription elongation factor A (SII), 2 |
| chr4_+_107367757 | 4.58 |
ENSMUST00000139560.1
|
Ndc1
|
NDC1 transmembrane nucleoporin |
| chr13_-_110280103 | 4.56 |
ENSMUST00000167824.1
|
Rab3c
|
RAB3C, member RAS oncogene family |
| chr2_-_26021679 | 4.55 |
ENSMUST00000036509.7
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr1_+_172482199 | 4.54 |
ENSMUST00000135267.1
ENSMUST00000052629.6 ENSMUST00000111235.2 |
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr17_-_23673557 | 4.48 |
ENSMUST00000115489.1
|
Thoc6
|
THO complex 6 homolog (Drosophila) |
| chr5_+_30711564 | 4.39 |
ENSMUST00000114729.1
|
Dpysl5
|
dihydropyrimidinase-like 5 |
| chr2_+_173021902 | 4.36 |
ENSMUST00000029014.9
|
Rbm38
|
RNA binding motif protein 38 |
| chr10_-_81496329 | 4.35 |
ENSMUST00000020463.7
|
Ncln
|
nicalin homolog (zebrafish) |
| chr5_+_30711849 | 4.34 |
ENSMUST00000088081.4
ENSMUST00000101442.3 |
Dpysl5
|
dihydropyrimidinase-like 5 |
| chr13_-_35906324 | 4.32 |
ENSMUST00000174230.1
ENSMUST00000171686.2 |
Rpp40
|
ribonuclease P 40 subunit |
| chr2_-_26021532 | 4.30 |
ENSMUST00000136750.1
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr8_-_92355764 | 4.28 |
ENSMUST00000180102.1
ENSMUST00000179421.1 ENSMUST00000179222.1 ENSMUST00000179029.1 |
4933436C20Rik
|
RIKEN cDNA 4933436C20 gene |
| chr14_-_65833963 | 4.27 |
ENSMUST00000022613.9
|
Esco2
|
establishment of cohesion 1 homolog 2 (S. cerevisiae) |
| chr10_+_77033260 | 4.24 |
ENSMUST00000136925.1
ENSMUST00000130703.1 |
Slc19a1
|
solute carrier family 19 (folate transporter), member 1 |
| chr19_+_36083696 | 4.20 |
ENSMUST00000025714.7
|
Rpp30
|
ribonuclease P/MRP 30 subunit |
| chr5_+_21424934 | 4.20 |
ENSMUST00000056045.4
|
Fam185a
|
family with sequence similarity 185, member A |
| chr2_-_32353247 | 4.11 |
ENSMUST00000078352.5
ENSMUST00000113352.2 ENSMUST00000113365.1 |
Dnm1
|
dynamin 1 |
| chr9_+_26999668 | 4.09 |
ENSMUST00000039161.8
|
Thyn1
|
thymocyte nuclear protein 1 |
| chr19_-_41802028 | 4.08 |
ENSMUST00000026150.8
ENSMUST00000177495.1 ENSMUST00000163265.1 |
Arhgap19
|
Rho GTPase activating protein 19 |
| chr4_+_134468320 | 4.07 |
ENSMUST00000030636.4
ENSMUST00000127279.1 ENSMUST00000105867.1 |
Stmn1
|
stathmin 1 |
| chr2_-_32353283 | 4.05 |
ENSMUST00000091089.5
ENSMUST00000113350.1 |
Dnm1
|
dynamin 1 |
| chr2_-_71055534 | 4.04 |
ENSMUST00000090849.5
ENSMUST00000100037.2 ENSMUST00000112186.2 |
Mettl8
|
methyltransferase like 8 |
| chr5_+_108132885 | 3.98 |
ENSMUST00000047677.7
|
Ccdc18
|
coiled-coil domain containing 18 |
| chr3_+_88081997 | 3.92 |
ENSMUST00000071812.5
|
Iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr10_+_77033212 | 3.92 |
ENSMUST00000105410.3
|
Slc19a1
|
solute carrier family 19 (folate transporter), member 1 |
| chr15_-_81729864 | 3.92 |
ENSMUST00000171115.1
ENSMUST00000170134.1 ENSMUST00000052374.5 |
Rangap1
|
RAN GTPase activating protein 1 |
| chr2_+_129100995 | 3.91 |
ENSMUST00000103205.4
ENSMUST00000028874.7 |
Polr1b
|
polymerase (RNA) I polypeptide B |
| chr10_+_77033304 | 3.90 |
ENSMUST00000132984.1
|
Slc19a1
|
solute carrier family 19 (folate transporter), member 1 |
| chr4_+_126556935 | 3.87 |
ENSMUST00000048391.8
|
Clspn
|
claspin |
| chr10_-_80918212 | 3.85 |
ENSMUST00000057623.7
ENSMUST00000179022.1 |
Lmnb2
|
lamin B2 |
| chr17_+_24632671 | 3.84 |
ENSMUST00000047611.2
|
Nthl1
|
nth (endonuclease III)-like 1 (E.coli) |
| chr8_+_124722139 | 3.81 |
ENSMUST00000034463.3
|
Arv1
|
ARV1 homolog (yeast) |
| chr7_+_13278778 | 3.79 |
ENSMUST00000098814.4
ENSMUST00000146998.1 ENSMUST00000185145.1 |
Lig1
|
ligase I, DNA, ATP-dependent |
| chr19_+_53329413 | 3.79 |
ENSMUST00000025998.7
|
Mxi1
|
Max interacting protein 1 |
| chr10_+_40883469 | 3.73 |
ENSMUST00000019975.7
|
Wasf1
|
WAS protein family, member 1 |
| chr10_-_128565827 | 3.72 |
ENSMUST00000131728.1
ENSMUST00000026425.6 |
Pa2g4
|
proliferation-associated 2G4 |
| chr9_+_78109188 | 3.71 |
ENSMUST00000118869.1
ENSMUST00000125615.1 |
Ick
|
intestinal cell kinase |
| chr3_+_88532314 | 3.66 |
ENSMUST00000172699.1
|
Mex3a
|
mex3 homolog A (C. elegans) |
| chr4_-_49521036 | 3.66 |
ENSMUST00000057829.3
|
Mrpl50
|
mitochondrial ribosomal protein L50 |
| chr10_+_40883819 | 3.63 |
ENSMUST00000105509.1
|
Wasf1
|
WAS protein family, member 1 |
| chr16_-_18811615 | 3.58 |
ENSMUST00000096990.3
|
Cdc45
|
cell division cycle 45 |
| chr7_+_24112314 | 3.58 |
ENSMUST00000120006.1
ENSMUST00000005413.3 |
Zfp112
|
zinc finger protein 112 |
| chr1_-_20820213 | 3.58 |
ENSMUST00000053266.9
|
Mcm3
|
minichromosome maintenance deficient 3 (S. cerevisiae) |
| chr1_+_175880775 | 3.57 |
ENSMUST00000039725.6
|
Exo1
|
exonuclease 1 |
| chr1_+_172481788 | 3.54 |
ENSMUST00000127052.1
|
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr17_-_23673825 | 3.54 |
ENSMUST00000115490.1
ENSMUST00000047436.4 ENSMUST00000138190.1 ENSMUST00000095579.4 |
Thoc6
|
THO complex 6 homolog (Drosophila) |
| chr17_-_35673517 | 3.51 |
ENSMUST00000162266.1
ENSMUST00000160734.1 ENSMUST00000159852.1 ENSMUST00000160039.1 |
Gtf2h4
|
general transcription factor II H, polypeptide 4 |
| chr1_+_181150926 | 3.48 |
ENSMUST00000134115.1
ENSMUST00000111059.1 |
Cnih4
|
cornichon homolog 4 (Drosophila) |
| chr11_+_116671658 | 3.44 |
ENSMUST00000106378.1
ENSMUST00000144049.1 |
1810032O08Rik
|
RIKEN cDNA 1810032O08 gene |
| chr8_+_105860634 | 3.42 |
ENSMUST00000008594.7
|
Nutf2
|
nuclear transport factor 2 |
| chr3_+_79629074 | 3.39 |
ENSMUST00000029388.8
|
4930579G24Rik
|
RIKEN cDNA 4930579G24 gene |
| chr12_+_40446050 | 3.38 |
ENSMUST00000037488.6
|
Dock4
|
dedicator of cytokinesis 4 |
| chr8_+_57488053 | 3.37 |
ENSMUST00000180690.1
|
2500002B13Rik
|
RIKEN cDNA 2500002B13 gene |
| chr8_+_110919916 | 3.36 |
ENSMUST00000117534.1
ENSMUST00000034197.4 |
St3gal2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| chr15_+_78428650 | 3.36 |
ENSMUST00000159771.1
|
Kctd17
|
potassium channel tetramerisation domain containing 17 |
| chr18_-_77767752 | 3.31 |
ENSMUST00000048192.7
|
Haus1
|
HAUS augmin-like complex, subunit 1 |
| chr5_+_110839973 | 3.27 |
ENSMUST00000066160.1
|
Chek2
|
checkpoint kinase 2 |
| chr6_-_112696604 | 3.20 |
ENSMUST00000113182.1
ENSMUST00000113180.1 ENSMUST00000068487.5 ENSMUST00000077088.4 |
Rad18
|
RAD18 homolog (S. cerevisiae) |
| chr14_-_99099701 | 3.19 |
ENSMUST00000042471.9
|
Dis3
|
DIS3 mitotic control homolog (S. cerevisiae) |
| chr10_-_81496313 | 3.18 |
ENSMUST00000118498.1
|
Ncln
|
nicalin homolog (zebrafish) |
| chr1_-_60098104 | 3.17 |
ENSMUST00000143342.1
|
Wdr12
|
WD repeat domain 12 |
| chr19_-_47050823 | 3.15 |
ENSMUST00000026032.5
|
Pcgf6
|
polycomb group ring finger 6 |
| chr11_-_103028204 | 3.13 |
ENSMUST00000155490.1
|
Dcakd
|
dephospho-CoA kinase domain containing |
| chr5_-_34660068 | 3.12 |
ENSMUST00000041364.9
|
Nop14
|
NOP14 nucleolar protein |
| chr9_+_81863744 | 3.08 |
ENSMUST00000057067.3
|
Mei4
|
meiosis-specific, MEI4 homolog (S. cerevisiae) |
| chr6_+_21985903 | 3.05 |
ENSMUST00000137437.1
ENSMUST00000115383.2 |
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr1_-_60098135 | 3.04 |
ENSMUST00000141417.1
ENSMUST00000122038.1 |
Wdr12
|
WD repeat domain 12 |
| chr14_-_54517353 | 3.02 |
ENSMUST00000023873.5
|
Prmt5
|
protein arginine N-methyltransferase 5 |
| chrX_-_162829379 | 3.01 |
ENSMUST00000041370.4
ENSMUST00000112316.2 ENSMUST00000112315.1 |
Txlng
|
taxilin gamma |
| chr1_-_60097893 | 3.01 |
ENSMUST00000027173.8
|
Wdr12
|
WD repeat domain 12 |
| chr2_+_152962485 | 3.01 |
ENSMUST00000099197.2
ENSMUST00000103155.3 |
Ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr2_+_119112793 | 2.92 |
ENSMUST00000140939.1
ENSMUST00000028795.3 |
Rad51
|
RAD51 homolog |
| chr11_-_77489666 | 2.88 |
ENSMUST00000037593.7
ENSMUST00000092892.3 |
Ankrd13b
|
ankyrin repeat domain 13b |
| chr9_+_106281061 | 2.85 |
ENSMUST00000072206.6
|
Poc1a
|
POC1 centriolar protein homolog A (Chlamydomonas) |
| chr10_+_80142295 | 2.83 |
ENSMUST00000003156.8
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr7_+_101663705 | 2.81 |
ENSMUST00000106998.1
|
Clpb
|
ClpB caseinolytic peptidase B |
| chr1_-_134955847 | 2.80 |
ENSMUST00000168381.1
|
Ppp1r12b
|
protein phosphatase 1, regulatory (inhibitor) subunit 12B |
| chr10_+_80142358 | 2.75 |
ENSMUST00000105366.1
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr13_+_112660739 | 2.69 |
ENSMUST00000052514.4
|
Slc38a9
|
solute carrier family 38, member 9 |
| chr2_+_71055731 | 2.67 |
ENSMUST00000154704.1
ENSMUST00000135357.1 ENSMUST00000064141.5 ENSMUST00000112159.2 ENSMUST00000102701.3 |
Dcaf17
|
DDB1 and CUL4 associated factor 17 |
| chr4_+_126556994 | 2.67 |
ENSMUST00000147675.1
|
Clspn
|
claspin |
| chr19_+_46056539 | 2.66 |
ENSMUST00000111899.1
ENSMUST00000099392.3 ENSMUST00000062322.4 |
Pprc1
|
peroxisome proliferative activated receptor, gamma, coactivator-related 1 |
| chr4_+_116807714 | 2.63 |
ENSMUST00000102699.1
ENSMUST00000130359.1 |
Mutyh
|
mutY homolog (E. coli) |
| chr3_+_85574109 | 2.62 |
ENSMUST00000127348.1
ENSMUST00000107672.1 ENSMUST00000107674.1 |
Pet112
|
PET112 homolog (S. cerevisiae) |
| chr11_-_78183551 | 2.62 |
ENSMUST00000102483.4
|
Rpl23a
|
ribosomal protein L23A |
| chrX_-_8252304 | 2.62 |
ENSMUST00000115594.1
|
Ftsj1
|
FtsJ homolog 1 (E. coli) |
| chrX_-_8252334 | 2.61 |
ENSMUST00000115595.1
ENSMUST00000033513.3 |
Ftsj1
|
FtsJ homolog 1 (E. coli) |
| chr9_+_21032038 | 2.58 |
ENSMUST00000019616.4
|
Icam5
|
intercellular adhesion molecule 5, telencephalin |
| chr16_-_18811972 | 2.57 |
ENSMUST00000000028.7
ENSMUST00000115585.1 |
Cdc45
|
cell division cycle 45 |
| chr15_-_9140374 | 2.56 |
ENSMUST00000096482.3
ENSMUST00000110585.2 |
Skp2
|
S-phase kinase-associated protein 2 (p45) |
| chr8_-_124721956 | 2.55 |
ENSMUST00000117624.1
ENSMUST00000041614.8 ENSMUST00000118134.1 |
Ttc13
|
tetratricopeptide repeat domain 13 |
| chr15_-_43869993 | 2.54 |
ENSMUST00000067469.4
|
Tmem74
|
transmembrane protein 74 |
| chr6_+_35177386 | 2.53 |
ENSMUST00000043815.9
|
Nup205
|
nucleoporin 205 |
| chr3_-_10440054 | 2.53 |
ENSMUST00000099223.4
ENSMUST00000029047.6 |
Snx16
|
sorting nexin 16 |
| chr5_+_33658567 | 2.48 |
ENSMUST00000114426.3
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr6_+_71909046 | 2.48 |
ENSMUST00000055296.8
|
Polr1a
|
polymerase (RNA) I polypeptide A |
| chr10_-_128626464 | 2.45 |
ENSMUST00000026420.5
|
Rps26
|
ribosomal protein S26 |
| chr6_+_86849488 | 2.44 |
ENSMUST00000089519.6
ENSMUST00000113668.1 |
Aak1
|
AP2 associated kinase 1 |
| chr2_-_77946180 | 2.41 |
ENSMUST00000111824.1
ENSMUST00000111819.1 ENSMUST00000128963.1 |
Cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr11_-_116168138 | 2.41 |
ENSMUST00000139020.1
ENSMUST00000103031.1 ENSMUST00000124828.1 |
Fbf1
|
Fas (TNFRSF6) binding factor 1 |
| chr1_-_128102412 | 2.41 |
ENSMUST00000112538.1
ENSMUST00000086614.5 |
Zranb3
|
zinc finger, RAN-binding domain containing 3 |
| chr7_+_100227311 | 2.39 |
ENSMUST00000084935.3
|
Pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr2_+_167062934 | 2.37 |
ENSMUST00000125674.1
|
1500012F01Rik
|
RIKEN cDNA 1500012F01 gene |
| chr3_+_97158767 | 2.37 |
ENSMUST00000090759.4
|
Acp6
|
acid phosphatase 6, lysophosphatidic |
| chr8_-_70897407 | 2.37 |
ENSMUST00000054220.8
|
Rpl18a
|
ribosomal protein L18A |
| chr11_-_100712429 | 2.35 |
ENSMUST00000006973.5
ENSMUST00000103118.3 |
Kat2a
|
K(lysine) acetyltransferase 2A |
| chr15_+_76351303 | 2.34 |
ENSMUST00000023212.8
ENSMUST00000160172.1 |
Maf1
|
MAF1 homolog (S. cerevisiae) |
| chr11_-_4704334 | 2.34 |
ENSMUST00000058407.5
|
Uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr11_+_16951371 | 2.33 |
ENSMUST00000109635.1
ENSMUST00000061327.1 |
Fbxo48
|
F-box protein 48 |
| chr9_-_98601642 | 2.31 |
ENSMUST00000035034.8
|
Mrps22
|
mitochondrial ribosomal protein S22 |
| chr7_+_101663633 | 2.30 |
ENSMUST00000001884.7
|
Clpb
|
ClpB caseinolytic peptidase B |
| chr8_-_106337987 | 2.29 |
ENSMUST00000067512.7
|
Smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr1_+_54250673 | 2.28 |
ENSMUST00000027128.4
|
Ccdc150
|
coiled-coil domain containing 150 |
| chr7_-_45830776 | 2.28 |
ENSMUST00000107723.2
ENSMUST00000131384.1 |
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr18_-_80469664 | 2.27 |
ENSMUST00000036229.6
|
Ctdp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr12_+_79297345 | 2.26 |
ENSMUST00000079533.5
ENSMUST00000171210.1 |
Rad51b
|
RAD51 homolog B |
| chr17_-_35673738 | 2.25 |
ENSMUST00000001565.8
|
Gtf2h4
|
general transcription factor II H, polypeptide 4 |
| chr15_-_53902472 | 2.25 |
ENSMUST00000078673.6
|
Samd12
|
sterile alpha motif domain containing 12 |
| chr7_+_100227638 | 2.24 |
ENSMUST00000054436.8
|
Pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr7_-_44869788 | 2.23 |
ENSMUST00000046575.9
|
Ptov1
|
prostate tumor over expressed gene 1 |
| chr8_-_105851981 | 2.20 |
ENSMUST00000040776.4
|
Cenpt
|
centromere protein T |
| chr7_-_30559600 | 2.19 |
ENSMUST00000043975.4
ENSMUST00000156241.1 |
Lin37
|
lin-37 homolog (C. elegans) |
| chr3_-_87885558 | 2.18 |
ENSMUST00000005015.8
|
Prcc
|
papillary renal cell carcinoma (translocation-associated) |
| chr17_-_25727364 | 2.18 |
ENSMUST00000170070.1
ENSMUST00000048054.7 |
Chtf18
|
CTF18, chromosome transmission fidelity factor 18 |
| chr11_+_70970467 | 2.18 |
ENSMUST00000178822.1
ENSMUST00000108529.3 ENSMUST00000169965.1 ENSMUST00000167509.1 |
Rpain
|
RPA interacting protein |
| chr5_-_135934590 | 2.14 |
ENSMUST00000055808.5
|
Ywhag
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide |
| chr10_+_128015157 | 2.11 |
ENSMUST00000178041.1
ENSMUST00000026461.7 |
Prim1
|
DNA primase, p49 subunit |
| chr8_-_92356103 | 2.10 |
ENSMUST00000034183.3
|
4933436C20Rik
|
RIKEN cDNA 4933436C20 gene |
| chr7_-_30559828 | 2.08 |
ENSMUST00000108164.1
|
Lin37
|
lin-37 homolog (C. elegans) |
| chr15_-_76639840 | 2.07 |
ENSMUST00000166974.1
ENSMUST00000168185.1 |
Tonsl
|
tonsoku-like, DNA repair protein |
| chr8_-_57487801 | 2.07 |
ENSMUST00000034022.3
|
Sap30
|
sin3 associated polypeptide |
| chr4_+_116708624 | 2.03 |
ENSMUST00000106463.1
|
Ccdc163
|
coiled-coil domain containing 163 |
| chr8_-_72443772 | 2.02 |
ENSMUST00000019876.5
|
Calr3
|
calreticulin 3 |
| chr8_-_105707933 | 2.01 |
ENSMUST00000013299.9
|
Enkd1
|
enkurin domain containing 1 |
| chr4_+_149104130 | 1.97 |
ENSMUST00000103216.3
ENSMUST00000030816.3 |
Dffa
|
DNA fragmentation factor, alpha subunit |
| chr11_+_58307122 | 1.95 |
ENSMUST00000049353.8
|
Zfp692
|
zinc finger protein 692 |
| chr15_+_76904070 | 1.92 |
ENSMUST00000004072.8
|
Rpl8
|
ribosomal protein L8 |
| chr17_+_84626458 | 1.92 |
ENSMUST00000025101.8
|
Dync2li1
|
dynein cytoplasmic 2 light intermediate chain 1 |
| chr11_-_115627948 | 1.92 |
ENSMUST00000154623.1
ENSMUST00000106503.3 ENSMUST00000141614.1 |
Slc25a19
|
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr1_-_134955908 | 1.91 |
ENSMUST00000045665.6
ENSMUST00000086444.4 ENSMUST00000112163.1 |
Ppp1r12b
|
protein phosphatase 1, regulatory (inhibitor) subunit 12B |
| chr6_+_35177610 | 1.91 |
ENSMUST00000170234.1
|
Nup205
|
nucleoporin 205 |
| chr4_-_139352298 | 1.89 |
ENSMUST00000030513.6
ENSMUST00000155257.1 |
Mrto4
|
MRT4, mRNA turnover 4, homolog (S. cerevisiae) |
| chr2_+_32095518 | 1.87 |
ENSMUST00000057423.5
|
Ppapdc3
|
phosphatidic acid phosphatase type 2 domain containing 3 |
| chr13_-_54468805 | 1.86 |
ENSMUST00000026990.5
|
Thoc3
|
THO complex 3 |
| chr11_+_98907801 | 1.85 |
ENSMUST00000092706.6
|
Cdc6
|
cell division cycle 6 |
| chr7_-_13054514 | 1.85 |
ENSMUST00000182087.1
|
Mzf1
|
myeloid zinc finger 1 |
| chr13_-_86046901 | 1.85 |
ENSMUST00000131011.1
|
Cox7c
|
cytochrome c oxidase subunit VIIc |
| chr4_-_139352538 | 1.84 |
ENSMUST00000102503.3
|
Mrto4
|
MRT4, mRNA turnover 4, homolog (S. cerevisiae) |
| chr11_-_70015346 | 1.83 |
ENSMUST00000018718.7
ENSMUST00000102574.3 |
Acadvl
|
acyl-Coenzyme A dehydrogenase, very long chain |
| chr2_-_148732457 | 1.82 |
ENSMUST00000028926.6
|
Napb
|
N-ethylmaleimide sensitive fusion protein attachment protein beta |
| chr11_-_50827681 | 1.82 |
ENSMUST00000109135.2
|
Zfp354c
|
zinc finger protein 354C |
| chr2_-_77946375 | 1.82 |
ENSMUST00000065889.3
|
Cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr4_-_41503046 | 1.82 |
ENSMUST00000054920.4
|
AI464131
|
expressed sequence AI464131 |
| chr9_+_83548309 | 1.81 |
ENSMUST00000113215.3
|
Sh3bgrl2
|
SH3 domain binding glutamic acid-rich protein like 2 |
| chr5_+_33658550 | 1.80 |
ENSMUST00000152847.1
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr1_+_178187721 | 1.80 |
ENSMUST00000159284.1
|
Desi2
|
desumoylating isopeptidase 2 |
| chr7_-_99483645 | 1.75 |
ENSMUST00000107096.1
ENSMUST00000032998.6 |
Rps3
|
ribosomal protein S3 |
| chr4_-_56947411 | 1.74 |
ENSMUST00000107609.3
ENSMUST00000068792.6 |
Tmem245
|
transmembrane protein 245 |
| chr6_-_120357342 | 1.74 |
ENSMUST00000163827.1
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr2_-_31845925 | 1.74 |
ENSMUST00000028188.7
|
Fibcd1
|
fibrinogen C domain containing 1 |
| chr11_-_115628125 | 1.74 |
ENSMUST00000155709.1
ENSMUST00000021089.4 |
Slc25a19
|
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr1_+_179803376 | 1.74 |
ENSMUST00000097454.2
|
Gm10518
|
predicted gene 10518 |
| chr4_+_49521176 | 1.73 |
ENSMUST00000042964.6
ENSMUST00000107696.1 |
Zfp189
|
zinc finger protein 189 |
| chr15_+_76351288 | 1.73 |
ENSMUST00000161527.1
ENSMUST00000160853.1 |
Maf1
|
MAF1 homolog (S. cerevisiae) |
| chr5_-_136135989 | 1.72 |
ENSMUST00000150406.1
ENSMUST00000006301.4 |
Lrwd1
|
leucine-rich repeats and WD repeat domain containing 1 |
| chr1_-_16656843 | 1.72 |
ENSMUST00000115352.3
|
Tceb1
|
transcription elongation factor B (SIII), polypeptide 1 |
| chr11_+_70970181 | 1.71 |
ENSMUST00000018593.3
|
Rpain
|
RPA interacting protein |
| chr3_+_89183131 | 1.70 |
ENSMUST00000140473.1
ENSMUST00000041913.6 |
Fam189b
|
family with sequence similarity 189, member B |
| chr11_-_115628260 | 1.69 |
ENSMUST00000178003.1
|
Slc25a19
|
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr2_-_77946331 | 1.68 |
ENSMUST00000111821.2
ENSMUST00000111818.1 |
Cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hcfc1_Six5_Smarcc2_Zfp143
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.5 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 2.0 | 6.1 | GO:0071163 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 2.0 | 6.1 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 2.0 | 12.1 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 1.9 | 13.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 1.6 | 10.9 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 1.3 | 5.3 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 1.3 | 3.8 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 1.2 | 3.7 | GO:0036292 | DNA rewinding(GO:0036292) |
| 1.2 | 8.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 1.1 | 3.4 | GO:1904046 | protein localization to nuclear pore(GO:0090204) negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 1.1 | 5.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 1.1 | 3.3 | GO:0051325 | interphase(GO:0051325) mitotic interphase(GO:0051329) |
| 1.1 | 7.4 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 1.0 | 3.8 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.9 | 3.7 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.9 | 9.9 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.9 | 10.5 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.9 | 4.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.8 | 0.8 | GO:0032240 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.8 | 30.7 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.8 | 3.2 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.8 | 2.4 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.8 | 10.0 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.7 | 9.6 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.7 | 4.2 | GO:0035247 | peptidyl-arginine omega-N-methylation(GO:0035247) |
| 0.7 | 1.4 | GO:0051106 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.7 | 2.0 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.6 | 1.9 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.6 | 3.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.6 | 6.0 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.6 | 6.5 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.6 | 4.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.6 | 1.7 | GO:0021933 | radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.6 | 1.7 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.5 | 3.8 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.5 | 2.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.5 | 2.5 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.5 | 3.9 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.5 | 1.0 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.5 | 1.4 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.5 | 2.8 | GO:0010825 | growth plate cartilage chondrocyte development(GO:0003431) positive regulation of centrosome duplication(GO:0010825) |
| 0.5 | 2.7 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.4 | 2.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.4 | 1.7 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.4 | 4.0 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.4 | 4.4 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.4 | 1.6 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.4 | 1.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.4 | 1.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.4 | 2.7 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.4 | 1.5 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.4 | 2.3 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.4 | 1.1 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.4 | 1.1 | GO:1904023 | regulation of fermentation(GO:0043465) regulation of NAD metabolic process(GO:1902688) regulation of glucose catabolic process to lactate via pyruvate(GO:1904023) |
| 0.3 | 2.4 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.3 | 2.0 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.3 | 1.0 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.3 | 1.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.3 | 1.0 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.3 | 2.9 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.3 | 0.3 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.3 | 1.3 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.3 | 1.6 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.3 | 1.0 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.3 | 3.5 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.3 | 3.8 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.3 | 2.2 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.3 | 10.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.3 | 1.2 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.3 | 1.5 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.3 | 2.7 | GO:0032070 | regulation of deoxyribonuclease activity(GO:0032070) |
| 0.3 | 1.5 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.3 | 4.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.3 | 0.3 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.3 | 3.9 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.3 | 1.7 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 2.2 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.3 | 5.5 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.3 | 1.4 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.3 | 8.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.3 | 4.9 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.3 | 4.2 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.3 | 1.0 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.2 | 1.0 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.2 | 1.5 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.2 | 0.5 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.2 | 3.9 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.2 | 5.8 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.2 | 1.2 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.2 | 0.7 | GO:2000556 | regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) |
| 0.2 | 0.7 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.2 | 1.4 | GO:1904923 | regulation of mitophagy in response to mitochondrial depolarization(GO:1904923) |
| 0.2 | 0.5 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.2 | 2.0 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.2 | 2.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 | 0.2 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.2 | 2.0 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.2 | 1.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 16.1 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.2 | 1.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.2 | 2.6 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 2.3 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.2 | 2.5 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.2 | 0.2 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.2 | 0.6 | GO:1990927 | calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.2 | 7.0 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.2 | 6.2 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 0.6 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.2 | 0.4 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.2 | 0.8 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.2 | 1.4 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.2 | 1.1 | GO:2000259 | positive regulation of complement activation(GO:0045917) progesterone receptor signaling pathway(GO:0050847) positive regulation of protein activation cascade(GO:2000259) |
| 0.2 | 0.6 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.2 | 0.4 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.2 | 0.5 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.2 | 1.2 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.2 | 0.7 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.2 | 0.3 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.2 | 0.5 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.2 | 6.2 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.2 | 2.0 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.2 | 1.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 0.7 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.2 | 0.5 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.2 | 0.8 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.2 | 3.1 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.2 | 0.3 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.2 | 1.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 0.5 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.2 | 0.5 | GO:0002276 | basophil activation involved in immune response(GO:0002276) basophil activation(GO:0045575) |
| 0.2 | 0.6 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.2 | 1.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 2.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 1.5 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 3.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 0.9 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.1 | 4.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 1.4 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 1.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 2.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.6 | GO:0035482 | negative regulation of luteinizing hormone secretion(GO:0033685) gastric motility(GO:0035482) |
| 0.1 | 0.6 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.1 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) |
| 0.1 | 3.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 9.0 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 1.6 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.3 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 0.8 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.6 | GO:0034351 | negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.1 | 0.9 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.2 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.1 | 0.2 | GO:0031126 | snoRNA 3'-end processing(GO:0031126) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 1.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 1.7 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 0.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 1.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 3.8 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 1.1 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.1 | 3.9 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.1 | 0.7 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 1.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 5.1 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 0.5 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 1.4 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 1.5 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 0.6 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 0.5 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.1 | 0.5 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.3 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 1.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 1.1 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.1 | 0.6 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.1 | 0.6 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.6 | GO:1904851 | positive regulation of establishment of protein localization to telomere(GO:1904851) |
| 0.1 | 0.4 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 1.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.5 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.1 | 0.2 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.1 | 0.7 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 1.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 1.0 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.2 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.1 | 0.8 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 4.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 2.7 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.3 | GO:0030647 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.1 | 0.5 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.1 | 1.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 1.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 1.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 0.3 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.1 | 1.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 4.7 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.1 | 0.4 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.1 | 0.9 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 1.7 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.1 | 0.5 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 0.5 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 0.3 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 3.2 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 0.1 | GO:0002362 | CD4-positive, CD25-positive, alpha-beta regulatory T cell lineage commitment(GO:0002362) |
| 0.1 | 0.3 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 3.3 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.6 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 7.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.5 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 1.3 | GO:0052646 | alditol phosphate metabolic process(GO:0052646) |
| 0.1 | 0.2 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 1.2 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.1 | 0.3 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 0.3 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.2 | GO:0035928 | RNA import into mitochondrion(GO:0035927) rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 0.2 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.1 | 0.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.2 | GO:0052472 | modulation by host of viral transcription(GO:0043921) modulation of transcription in other organism involved in symbiotic interaction(GO:0052312) modulation by host of symbiont transcription(GO:0052472) |
| 0.1 | 0.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.1 | 0.2 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 1.1 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.1 | 0.8 | GO:0031034 | myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 2.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 1.9 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 0.2 | GO:0090343 | positive regulation of cell aging(GO:0090343) positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.2 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.2 | GO:0042699 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 0.2 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.2 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.1 | 0.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.6 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.2 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 5.7 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.1 | 0.6 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.2 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 0.1 | GO:0070350 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.0 | 0.7 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.4 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.4 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 0.4 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 3.1 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 10.2 | GO:0000375 | RNA splicing, via transesterification reactions(GO:0000375) |
| 0.0 | 1.1 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.0 | 0.7 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.4 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 3.0 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.2 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 2.0 | GO:0043297 | apical junction assembly(GO:0043297) |
| 0.0 | 0.3 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.6 | GO:0002176 | male germ cell proliferation(GO:0002176) |
| 0.0 | 0.4 | GO:0034145 | Toll signaling pathway(GO:0008063) positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 0.3 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 1.8 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.6 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.0 | 0.8 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 1.4 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.7 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.4 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 1.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0035822 | meiotic gene conversion(GO:0006311) regulation of reciprocal meiotic recombination(GO:0010520) positive regulation of reciprocal meiotic recombination(GO:0010845) gene conversion(GO:0035822) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 3.1 | GO:0009411 | response to UV(GO:0009411) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 8.0 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 1.4 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.1 | GO:0015822 | mitochondrial ornithine transport(GO:0000066) ornithine transport(GO:0015822) |
| 0.0 | 0.3 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.0 | 0.3 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.4 | GO:1901898 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.2 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 3.1 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.1 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.8 | GO:0006401 | RNA catabolic process(GO:0006401) |
| 0.0 | 0.3 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.4 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.8 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.8 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.4 | GO:1901992 | positive regulation of mitotic cell cycle phase transition(GO:1901992) |
| 0.0 | 0.1 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.2 | GO:0055119 | relaxation of cardiac muscle(GO:0055119) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.3 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.0 | 0.1 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.2 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:0015755 | fructose transport(GO:0015755) |
| 0.0 | 1.8 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.1 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.0 | 0.2 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 1.2 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 1.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.6 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.0 | 0.4 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.8 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) |
| 0.0 | 0.6 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.1 | GO:0018202 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) peptidyl-histidine modification(GO:0018202) |
| 0.0 | 1.4 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 0.2 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.1 | GO:0023021 | termination of signal transduction(GO:0023021) termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 0.3 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 7.4 | GO:0051301 | cell division(GO:0051301) |
| 0.0 | 0.1 | GO:1903874 | ferrous iron transmembrane transport(GO:1903874) |
| 0.0 | 7.3 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 0.2 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.1 | GO:1900225 | regulation of NLRP3 inflammasome complex assembly(GO:1900225) negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.2 | GO:0035825 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.3 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.5 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 2.3 | GO:0048167 | regulation of synaptic plasticity(GO:0048167) |
| 0.0 | 0.2 | GO:0009251 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.3 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.4 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.5 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.1 | GO:0021772 | olfactory bulb development(GO:0021772) |
| 0.0 | 0.1 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.0 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.1 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.7 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.0 | 0.2 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.0 | 0.4 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.0 | 0.7 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.1 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.2 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.2 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.6 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.0 | 0.1 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.3 | GO:2001022 | positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.0 | 0.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.0 | GO:1903181 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.0 | 1.0 | GO:0010564 | regulation of cell cycle process(GO:0010564) |
| 0.0 | 0.1 | GO:0001709 | cell fate determination(GO:0001709) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 18.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 2.0 | 6.1 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 1.9 | 15.5 | GO:0005818 | aster(GO:0005818) |
| 1.6 | 1.6 | GO:0070552 | BRISC complex(GO:0070552) |
| 1.4 | 5.8 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 1.4 | 9.6 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 1.3 | 3.9 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 1.2 | 5.9 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 1.1 | 6.3 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 1.0 | 10.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 1.0 | 9.9 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.9 | 4.4 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.8 | 10.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.8 | 19.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.7 | 5.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.7 | 5.6 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.7 | 2.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.6 | 3.2 | GO:0030689 | Noc complex(GO:0030689) |
| 0.6 | 5.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.6 | 1.7 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.6 | 3.9 | GO:0005638 | lamin filament(GO:0005638) |
| 0.5 | 4.9 | GO:0000801 | central element(GO:0000801) |
| 0.5 | 6.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.5 | 2.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.5 | 2.7 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.5 | 7.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.4 | 3.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.4 | 2.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.4 | 8.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.4 | 1.2 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.4 | 1.2 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.4 | 3.8 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.4 | 0.7 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.4 | 1.8 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.4 | 1.8 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.3 | 13.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 3.7 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.3 | 1.0 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.3 | 3.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.3 | 7.5 | GO:0001741 | XY body(GO:0001741) |
| 0.3 | 1.8 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.3 | 3.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.3 | 1.2 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.3 | 2.3 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.3 | 0.9 | GO:0055087 | Ski complex(GO:0055087) |
| 0.3 | 3.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.3 | 0.8 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.3 | 0.8 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.3 | 3.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 2.7 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.3 | 3.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.3 | 1.9 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.3 | 2.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.3 | 2.8 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.3 | 1.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 2.7 | GO:0035859 | Seh1-associated complex(GO:0035859) GATOR2 complex(GO:0061700) |
| 0.2 | 2.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.2 | 1.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.2 | 0.7 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.2 | 6.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.2 | 1.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 7.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 12.3 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 0.6 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.2 | 1.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.2 | 0.8 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 0.2 | 2.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.2 | 2.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 1.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.2 | 0.9 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.2 | 0.5 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 5.3 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.2 | 2.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.2 | 1.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 3.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 1.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.2 | 1.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 2.2 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.2 | 1.7 | GO:0000796 | condensin complex(GO:0000796) |
| 0.2 | 8.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 2.0 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.2 | 4.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.7 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 0.6 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 2.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 2.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 1.8 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.7 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 9.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.4 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 17.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 0.9 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 3.8 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 1.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 1.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 2.1 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 1.0 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.7 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 1.0 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 5.8 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 0.6 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 1.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 0.3 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 1.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 7.7 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 1.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.0 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 10.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.3 | GO:1990047 | spindle matrix(GO:1990047) |
| 0.1 | 0.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 1.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 6.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 2.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) small ribosomal subunit(GO:0015935) |
| 0.1 | 1.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 1.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 1.0 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.1 | 4.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 0.2 | GO:0031533 | mRNA cap methyltransferase complex(GO:0031533) |
| 0.1 | 2.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.9 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.5 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 0.7 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 2.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 2.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 1.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 3.2 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 2.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 1.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 0.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 6.1 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 4.3 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 2.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 4.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.6 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 0.3 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.9 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 2.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.9 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.5 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.2 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.2 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.3 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.6 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 3.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.8 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 28.7 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 1.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.0 | 1.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 4.8 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.9 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.7 | GO:0032155 | cell division site(GO:0032153) cleavage furrow(GO:0032154) cell division site part(GO:0032155) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 1.6 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 6.7 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 1.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 1.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.0 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 1.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.6 | GO:0098800 | inner mitochondrial membrane protein complex(GO:0098800) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.3 | GO:0032590 | dendrite membrane(GO:0032590) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 12.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 1.6 | 14.0 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 1.4 | 9.5 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 1.3 | 7.9 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 1.1 | 5.6 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 1.0 | 3.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.9 | 4.7 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.9 | 9.7 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.8 | 3.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.8 | 3.3 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) |
| 0.8 | 8.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.8 | 4.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.8 | 5.3 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.7 | 5.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.7 | 4.6 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.6 | 8.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.6 | 2.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.6 | 4.0 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.6 | 3.9 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.6 | 4.5 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.5 | 24.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.5 | 2.6 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.5 | 1.5 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.5 | 1.5 | GO:0008988 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.5 | 7.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.5 | 5.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.5 | 1.4 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.5 | 1.8 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.5 | 3.2 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.4 | 4.3 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.4 | 3.8 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.4 | 1.6 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.4 | 3.0 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.4 | 1.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.3 | 3.5 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.3 | 8.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.3 | 5.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.3 | 5.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.3 | 8.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.3 | 0.9 | GO:0071209 | histone pre-mRNA DCP binding(GO:0071208) U7 snRNA binding(GO:0071209) |
| 0.3 | 1.5 | GO:0008061 | chitin binding(GO:0008061) |
| 0.3 | 2.3 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.3 | 2.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.3 | 3.0 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.3 | 2.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.3 | 2.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.3 | 2.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.3 | 1.3 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.2 | 1.0 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.2 | 7.7 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.2 | 2.4 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 0.9 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.2 | 0.2 | GO:0031762 | alpha-1A adrenergic receptor binding(GO:0031691) alpha-1B adrenergic receptor binding(GO:0031692) follicle-stimulating hormone receptor binding(GO:0031762) |
| 0.2 | 0.8 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 0.6 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 5.8 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.2 | 1.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 1.6 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.2 | 2.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 1.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.2 | 4.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 4.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 0.6 | GO:0043404 | corticotrophin-releasing factor receptor activity(GO:0015056) corticotropin-releasing hormone receptor activity(GO:0043404) |
| 0.2 | 0.2 | GO:0032135 | DNA insertion or deletion binding(GO:0032135) |
| 0.2 | 0.7 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.2 | 0.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 1.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 1.4 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.2 | 1.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 1.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 2.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 3.6 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 24.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.4 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.1 | 0.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 2.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.4 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 8.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 1.0 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 0.4 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.1 | 1.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.5 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.4 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 1.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 0.8 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 8.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 0.8 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 2.7 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.2 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.1 | 0.6 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 1.5 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 0.8 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 0.6 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.0 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 0.3 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.1 | 1.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 3.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 1.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 2.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 1.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.4 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.1 | 0.7 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 0.9 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 1.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.5 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 0.6 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.8 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.3 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.1 | 0.4 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.7 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 0.3 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.1 | 0.3 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.1 | 0.5 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.1 | 0.9 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 2.1 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 2.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 3.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 0.5 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 0.2 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.1 | 1.9 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 0.4 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 6.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.3 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 2.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 0.3 | GO:0035614 | U1 snRNA binding(GO:0030619) snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 5.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 1.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 2.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 1.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.3 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.4 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 2.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.9 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 2.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 1.0 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 3.6 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.7 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.7 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.8 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.9 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 10.9 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.8 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 2.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.4 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 1.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.6 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 6.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 1.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.4 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 0.0 | 2.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.2 | GO:0016859 | cis-trans isomerase activity(GO:0016859) |
| 0.0 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.0 | 44.1 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.6 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.9 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.4 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.4 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 1.1 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 1.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.2 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 2.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 2.0 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 1.2 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 0.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.1 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.5 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.5 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 1.2 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.0 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 1.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 7.4 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 1.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 2.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.4 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.3 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 47.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.6 | 23.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.3 | 0.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.3 | 13.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 3.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 6.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.2 | 8.8 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.2 | 5.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 6.1 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 16.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 6.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 3.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.6 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 0.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 4.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 0.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 0.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 0.8 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 0.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 2.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.9 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.3 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 2.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.0 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.9 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.1 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 16.9 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.6 | 9.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.6 | 12.3 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.6 | 9.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.5 | 3.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.5 | 1.6 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.5 | 4.6 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.5 | 12.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.4 | 1.3 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.4 | 8.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.4 | 2.1 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.4 | 6.9 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.4 | 7.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.4 | 36.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.3 | 7.9 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.2 | 5.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.2 | 3.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 0.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 19.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.2 | 3.2 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.2 | 1.6 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.2 | 3.7 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.2 | 2.8 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 9.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 2.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 3.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 1.2 | REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | Genes involved in Formation of RNA Pol II elongation complex |
| 0.1 | 0.2 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 2.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 5.7 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 1.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 2.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 1.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.7 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 2.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.1 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 2.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.9 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 2.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 1.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 1.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.9 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.6 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 1.9 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.5 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 2.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 3.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.9 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 4.4 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 1.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.6 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.0 | 0.4 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.9 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.4 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |