Project
avrg: 12D miR HR13_24
Navigation
Downloads
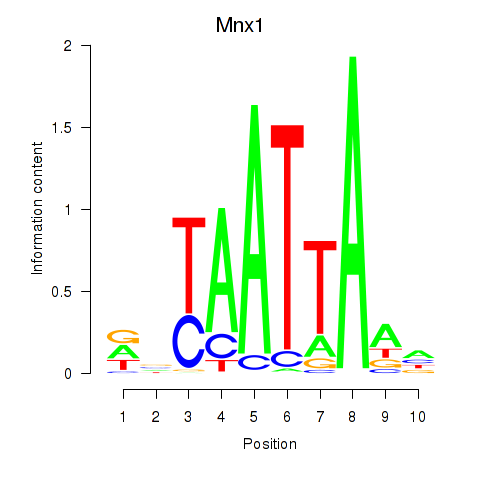
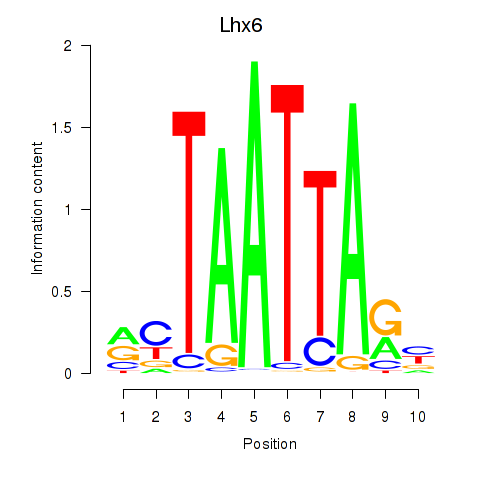
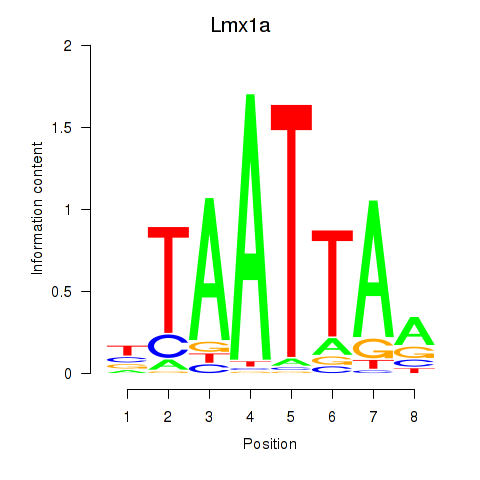
Results for Mnx1_Lhx6_Lmx1a
Z-value: 2.41
Transcription factors associated with Mnx1_Lhx6_Lmx1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mnx1
|
ENSMUSG00000001566.8 | motor neuron and pancreas homeobox 1 |
|
Lhx6
|
ENSMUSG00000026890.13 | LIM homeobox protein 6 |
|
Lmx1a
|
ENSMUSG00000026686.8 | LIM homeobox transcription factor 1 alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mnx1 | mm10_v2_chr5_-_29478470_29478597 | 0.87 | 5.4e-04 | Click! |
| Lmx1a | mm10_v2_chr1_+_167689552_167689563 | 0.68 | 2.1e-02 | Click! |
| Lhx6 | mm10_v2_chr2_-_36104060_36104073 | 0.66 | 2.7e-02 | Click! |
Activity profile of Mnx1_Lhx6_Lmx1a motif
Sorted Z-values of Mnx1_Lhx6_Lmx1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_25372315 | 4.48 |
ENSMUST00000028329.6
ENSMUST00000114293.2 ENSMUST00000100323.2 |
Sapcd2
|
suppressor APC domain containing 2 |
| chr10_+_4611971 | 4.19 |
ENSMUST00000105590.1
ENSMUST00000067086.7 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr3_-_49757257 | 4.19 |
ENSMUST00000035931.7
|
Pcdh18
|
protocadherin 18 |
| chr9_+_65890237 | 4.02 |
ENSMUST00000045802.6
|
2810417H13Rik
|
RIKEN cDNA 2810417H13 gene |
| chr18_+_23415400 | 3.99 |
ENSMUST00000115832.2
ENSMUST00000047954.7 |
Dtna
|
dystrobrevin alpha |
| chr16_-_45724600 | 3.86 |
ENSMUST00000096057.4
|
Tagln3
|
transgelin 3 |
| chr9_+_32116040 | 3.42 |
ENSMUST00000174641.1
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr15_-_79285502 | 3.13 |
ENSMUST00000165408.1
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr9_+_72806874 | 2.90 |
ENSMUST00000055535.8
|
Prtg
|
protogenin homolog (Gallus gallus) |
| chr15_-_79285470 | 2.66 |
ENSMUST00000170955.1
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr3_-_88410295 | 2.66 |
ENSMUST00000056370.7
|
Pmf1
|
polyamine-modulated factor 1 |
| chrX_-_74246534 | 2.63 |
ENSMUST00000101454.2
ENSMUST00000033699.6 |
Flna
|
filamin, alpha |
| chr8_-_62123106 | 2.62 |
ENSMUST00000034052.6
ENSMUST00000034054.7 |
Anxa10
|
annexin A10 |
| chr5_+_104202609 | 2.56 |
ENSMUST00000066708.5
|
Dmp1
|
dentin matrix protein 1 |
| chr11_+_115334731 | 2.50 |
ENSMUST00000106543.1
ENSMUST00000019006.4 |
Otop3
|
otopetrin 3 |
| chr7_+_30493622 | 2.44 |
ENSMUST00000058280.6
ENSMUST00000133318.1 ENSMUST00000142575.1 ENSMUST00000131040.1 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr3_+_122044428 | 2.14 |
ENSMUST00000013995.8
|
Abca4
|
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr7_+_64501687 | 2.14 |
ENSMUST00000032732.8
|
Apba2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr19_-_55241236 | 1.87 |
ENSMUST00000069183.6
|
Gucy2g
|
guanylate cyclase 2g |
| chr3_+_54361103 | 1.85 |
ENSMUST00000107985.3
ENSMUST00000117373.1 ENSMUST00000073012.6 ENSMUST00000081564.6 |
Postn
|
periostin, osteoblast specific factor |
| chr6_+_145934113 | 1.81 |
ENSMUST00000032383.7
|
Sspn
|
sarcospan |
| chr7_-_45830776 | 1.73 |
ENSMUST00000107723.2
ENSMUST00000131384.1 |
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr10_+_88091070 | 1.69 |
ENSMUST00000048621.7
|
Pmch
|
pro-melanin-concentrating hormone |
| chr2_+_162987502 | 1.68 |
ENSMUST00000117123.1
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr2_+_162987330 | 1.67 |
ENSMUST00000018012.7
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr5_-_62766153 | 1.61 |
ENSMUST00000076623.4
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr12_+_38780284 | 1.60 |
ENSMUST00000162563.1
ENSMUST00000161164.1 ENSMUST00000160996.1 |
Etv1
|
ets variant gene 1 |
| chrX_+_169685191 | 1.59 |
ENSMUST00000112104.1
ENSMUST00000112107.1 |
Mid1
|
midline 1 |
| chr4_+_108719649 | 1.54 |
ENSMUST00000178992.1
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr5_+_118169712 | 1.49 |
ENSMUST00000054836.6
|
Hrk
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chrM_+_2743 | 1.49 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr11_-_87359011 | 1.46 |
ENSMUST00000055438.4
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing) |
| chr6_-_136781718 | 1.44 |
ENSMUST00000078095.6
ENSMUST00000032338.7 |
Gucy2c
|
guanylate cyclase 2c |
| chr1_-_158356258 | 1.43 |
ENSMUST00000004133.8
|
Brinp2
|
bone morphogenic protein/retinoic acid inducible neural-specific 2 |
| chr3_+_68572245 | 1.43 |
ENSMUST00000170788.2
|
Schip1
|
schwannomin interacting protein 1 |
| chr6_+_8948608 | 1.42 |
ENSMUST00000160300.1
|
Nxph1
|
neurexophilin 1 |
| chr12_+_38780817 | 1.41 |
ENSMUST00000160856.1
|
Etv1
|
ets variant gene 1 |
| chrX_+_166344692 | 1.36 |
ENSMUST00000112223.1
ENSMUST00000112224.1 ENSMUST00000112229.2 ENSMUST00000112228.1 ENSMUST00000112227.2 ENSMUST00000112226.2 |
Gpm6b
|
glycoprotein m6b |
| chr3_+_55782500 | 1.33 |
ENSMUST00000075422.4
|
Mab21l1
|
mab-21-like 1 (C. elegans) |
| chrX_+_134308084 | 1.33 |
ENSMUST00000081064.5
ENSMUST00000101251.1 ENSMUST00000129782.1 |
Cenpi
|
centromere protein I |
| chrX_-_134111852 | 1.32 |
ENSMUST00000033610.6
|
Nox1
|
NADPH oxidase 1 |
| chr18_+_4993795 | 1.26 |
ENSMUST00000153016.1
|
Svil
|
supervillin |
| chr8_-_31918203 | 1.23 |
ENSMUST00000073884.4
|
Nrg1
|
neuregulin 1 |
| chrX_+_150547375 | 1.23 |
ENSMUST00000066337.6
ENSMUST00000112715.1 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chrX_-_102157065 | 1.21 |
ENSMUST00000056904.2
|
Ercc6l
|
excision repair cross-complementing rodent repair deficiency complementation group 6 like |
| chr7_+_64502090 | 1.17 |
ENSMUST00000137732.1
|
Apba2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr16_-_63864114 | 1.16 |
ENSMUST00000064405.6
|
Epha3
|
Eph receptor A3 |
| chr5_-_138170992 | 1.12 |
ENSMUST00000139983.1
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr2_-_45117349 | 1.07 |
ENSMUST00000176438.2
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr6_+_34746368 | 1.04 |
ENSMUST00000142716.1
|
Cald1
|
caldesmon 1 |
| chr19_-_15924560 | 1.03 |
ENSMUST00000162053.1
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr11_-_87108656 | 1.02 |
ENSMUST00000051395.8
|
Prr11
|
proline rich 11 |
| chr16_-_92400067 | 1.02 |
ENSMUST00000023672.8
|
Rcan1
|
regulator of calcineurin 1 |
| chr6_-_136875794 | 1.01 |
ENSMUST00000032342.1
|
Mgp
|
matrix Gla protein |
| chr15_-_103215285 | 1.01 |
ENSMUST00000122182.1
ENSMUST00000108813.3 ENSMUST00000127191.1 |
Cbx5
|
chromobox 5 |
| chr11_+_115307155 | 1.01 |
ENSMUST00000055490.2
|
Otop2
|
otopetrin 2 |
| chr4_-_99654983 | 1.00 |
ENSMUST00000136525.1
|
Gm12688
|
predicted gene 12688 |
| chr5_-_84417359 | 1.00 |
ENSMUST00000113401.1
|
Epha5
|
Eph receptor A5 |
| chrX_-_150657392 | 1.00 |
ENSMUST00000151403.2
ENSMUST00000087253.4 ENSMUST00000112709.1 ENSMUST00000163969.1 ENSMUST00000087258.3 |
Tro
|
trophinin |
| chr3_-_33082004 | 0.98 |
ENSMUST00000108225.3
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr2_-_33086366 | 0.98 |
ENSMUST00000049618.2
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr11_+_23306910 | 0.98 |
ENSMUST00000137823.1
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr15_+_55307743 | 0.97 |
ENSMUST00000023053.5
ENSMUST00000110221.2 ENSMUST00000110217.3 |
Col14a1
|
collagen, type XIV, alpha 1 |
| chr5_+_87925579 | 0.96 |
ENSMUST00000001667.6
ENSMUST00000113267.1 |
Csn3
|
casein kappa |
| chr3_+_94372794 | 0.96 |
ENSMUST00000029795.3
|
Rorc
|
RAR-related orphan receptor gamma |
| chr2_+_119047129 | 0.96 |
ENSMUST00000153300.1
ENSMUST00000028799.5 |
Casc5
|
cancer susceptibility candidate 5 |
| chr1_+_53061637 | 0.94 |
ENSMUST00000027269.5
|
Mstn
|
myostatin |
| chr9_+_96258697 | 0.93 |
ENSMUST00000179416.1
|
Tfdp2
|
transcription factor Dp 2 |
| chr19_-_15924928 | 0.93 |
ENSMUST00000025542.3
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr3_-_116253467 | 0.92 |
ENSMUST00000090473.5
|
Gpr88
|
G-protein coupled receptor 88 |
| chr8_-_122460666 | 0.92 |
ENSMUST00000006762.5
|
Snai3
|
snail homolog 3 (Drosophila) |
| chr7_-_45103747 | 0.89 |
ENSMUST00000003512.7
|
Fcgrt
|
Fc receptor, IgG, alpha chain transporter |
| chr9_-_20959785 | 0.89 |
ENSMUST00000177754.1
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1 |
| chr6_+_63255971 | 0.88 |
ENSMUST00000159561.1
ENSMUST00000095852.3 |
Grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr6_+_71909046 | 0.87 |
ENSMUST00000055296.8
|
Polr1a
|
polymerase (RNA) I polypeptide A |
| chrM_+_9452 | 0.87 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr18_-_75697639 | 0.86 |
ENSMUST00000165559.1
|
Ctif
|
CBP80/20-dependent translation initiation factor |
| chr11_+_23306884 | 0.85 |
ENSMUST00000180046.1
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr2_-_28916412 | 0.85 |
ENSMUST00000050776.2
ENSMUST00000113849.1 |
Barhl1
|
BarH-like 1 (Drosophila) |
| chr15_+_92597104 | 0.85 |
ENSMUST00000035399.8
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr2_+_116067213 | 0.83 |
ENSMUST00000152412.1
|
G630016G05Rik
|
RIKEN cDNA G630016G05 gene |
| chr19_+_30232921 | 0.83 |
ENSMUST00000025797.5
|
Mbl2
|
mannose-binding lectin (protein C) 2 |
| chrX_-_150657366 | 0.81 |
ENSMUST00000148604.1
|
Tro
|
trophinin |
| chr7_-_116198487 | 0.80 |
ENSMUST00000181981.1
|
Plekha7
|
pleckstrin homology domain containing, family A member 7 |
| chr2_+_152754156 | 0.79 |
ENSMUST00000010020.5
|
Cox4i2
|
cytochrome c oxidase subunit IV isoform 2 |
| chr1_+_161494649 | 0.77 |
ENSMUST00000086084.1
|
Tnfsf18
|
tumor necrosis factor (ligand) superfamily, member 18 |
| chr3_-_130730375 | 0.76 |
ENSMUST00000079085.6
|
Rpl34
|
ribosomal protein L34 |
| chr7_-_38019505 | 0.76 |
ENSMUST00000085513.4
|
Uri1
|
URI1, prefoldin-like chaperone |
| chr6_+_15196949 | 0.75 |
ENSMUST00000151301.1
ENSMUST00000131414.1 ENSMUST00000140557.1 ENSMUST00000115469.1 |
Foxp2
|
forkhead box P2 |
| chrX_-_74246364 | 0.75 |
ENSMUST00000130007.1
|
Flna
|
filamin, alpha |
| chr1_+_63176818 | 0.75 |
ENSMUST00000129339.1
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr17_+_12119274 | 0.75 |
ENSMUST00000024594.2
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr17_-_48432723 | 0.74 |
ENSMUST00000046549.3
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chrM_+_7005 | 0.73 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr1_+_40515362 | 0.72 |
ENSMUST00000027237.5
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chr3_-_94436574 | 0.72 |
ENSMUST00000029787.4
|
Oaz3
|
ornithine decarboxylase antizyme 3 |
| chr2_+_181767040 | 0.72 |
ENSMUST00000108756.1
|
Myt1
|
myelin transcription factor 1 |
| chr17_-_45733843 | 0.71 |
ENSMUST00000178179.1
|
1600014C23Rik
|
RIKEN cDNA 1600014C23 gene |
| chr2_-_17460610 | 0.71 |
ENSMUST00000145492.1
|
Nebl
|
nebulette |
| chr1_-_172027269 | 0.70 |
ENSMUST00000027837.6
ENSMUST00000111264.1 |
Vangl2
|
vang-like 2 (van gogh, Drosophila) |
| chr18_-_24603464 | 0.70 |
ENSMUST00000154205.1
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr13_-_97747399 | 0.69 |
ENSMUST00000144993.1
|
5330416C01Rik
|
RIKEN cDNA 5330416C01 gene |
| chr2_+_181767283 | 0.69 |
ENSMUST00000108757.2
|
Myt1
|
myelin transcription factor 1 |
| chr1_-_63176653 | 0.69 |
ENSMUST00000027111.8
ENSMUST00000168099.2 |
Ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| chr11_+_116843278 | 0.68 |
ENSMUST00000106370.3
|
Mettl23
|
methyltransferase like 23 |
| chr19_+_44992127 | 0.68 |
ENSMUST00000179305.1
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr12_-_73047179 | 0.65 |
ENSMUST00000050029.7
|
Six1
|
sine oculis-related homeobox 1 |
| chr6_+_7555053 | 0.65 |
ENSMUST00000090679.2
ENSMUST00000184986.1 |
Tac1
|
tachykinin 1 |
| chr1_-_152625212 | 0.65 |
ENSMUST00000027760.7
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr12_+_38781093 | 0.64 |
ENSMUST00000161513.1
|
Etv1
|
ets variant gene 1 |
| chr7_-_46667375 | 0.63 |
ENSMUST00000107669.2
|
Tph1
|
tryptophan hydroxylase 1 |
| chr12_-_54986328 | 0.61 |
ENSMUST00000038926.6
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr5_+_135106881 | 0.61 |
ENSMUST00000005507.3
|
Mlxipl
|
MLX interacting protein-like |
| chr3_-_130730310 | 0.60 |
ENSMUST00000062601.7
|
Rpl34
|
ribosomal protein L34 |
| chr12_-_54986363 | 0.58 |
ENSMUST00000173433.1
ENSMUST00000173803.1 |
Baz1a
Gm20403
|
bromodomain adjacent to zinc finger domain 1A predicted gene 20403 |
| chr19_+_44493472 | 0.58 |
ENSMUST00000041163.4
|
Wnt8b
|
wingless related MMTV integration site 8b |
| chr2_-_28916668 | 0.58 |
ENSMUST00000113847.1
|
Barhl1
|
BarH-like 1 (Drosophila) |
| chr16_-_89818338 | 0.57 |
ENSMUST00000164263.2
|
Tiam1
|
T cell lymphoma invasion and metastasis 1 |
| chr5_+_87925624 | 0.55 |
ENSMUST00000113271.2
|
Csn3
|
casein kappa |
| chr8_-_84662841 | 0.55 |
ENSMUST00000060427.4
|
Ier2
|
immediate early response 2 |
| chrX_+_56779437 | 0.55 |
ENSMUST00000114773.3
|
Fhl1
|
four and a half LIM domains 1 |
| chr9_+_7571396 | 0.54 |
ENSMUST00000120900.1
ENSMUST00000093896.3 ENSMUST00000151853.1 ENSMUST00000152878.1 |
Mmp27
|
matrix metallopeptidase 27 |
| chr2_-_174346712 | 0.54 |
ENSMUST00000168292.1
|
Gm20721
|
predicted gene, 20721 |
| chr10_+_85386813 | 0.53 |
ENSMUST00000105307.1
ENSMUST00000020231.3 |
Btbd11
|
BTB (POZ) domain containing 11 |
| chr6_+_125552948 | 0.53 |
ENSMUST00000112254.1
ENSMUST00000112253.1 ENSMUST00000001995.7 |
Vwf
|
Von Willebrand factor homolog |
| chr18_+_24603952 | 0.53 |
ENSMUST00000025120.6
|
Elp2
|
elongator acetyltransferase complex subunit 2 |
| chr7_+_103550368 | 0.53 |
ENSMUST00000106888.1
|
Olfr613
|
olfactory receptor 613 |
| chr1_-_183147461 | 0.52 |
ENSMUST00000171366.1
|
Disp1
|
dispatched homolog 1 (Drosophila) |
| chr2_+_20737306 | 0.52 |
ENSMUST00000114606.1
ENSMUST00000114608.1 |
Etl4
|
enhancer trap locus 4 |
| chr2_+_65620829 | 0.52 |
ENSMUST00000028377.7
|
Scn2a1
|
sodium channel, voltage-gated, type II, alpha 1 |
| chr16_-_5013505 | 0.51 |
ENSMUST00000023191.10
ENSMUST00000090453.5 |
Rogdi
|
rogdi homolog (Drosophila) |
| chr3_-_87174657 | 0.51 |
ENSMUST00000159976.1
ENSMUST00000107618.2 |
Kirrel
|
kin of IRRE like (Drosophila) |
| chr9_+_113812547 | 0.51 |
ENSMUST00000166734.2
ENSMUST00000111838.2 ENSMUST00000163895.2 |
Clasp2
|
CLIP associating protein 2 |
| chr9_-_55919605 | 0.51 |
ENSMUST00000037408.8
|
Scaper
|
S phase cyclin A-associated protein in the ER |
| chr15_-_43869993 | 0.50 |
ENSMUST00000067469.4
|
Tmem74
|
transmembrane protein 74 |
| chr15_-_9140374 | 0.50 |
ENSMUST00000096482.3
ENSMUST00000110585.2 |
Skp2
|
S-phase kinase-associated protein 2 (p45) |
| chr3_+_68869563 | 0.50 |
ENSMUST00000054551.2
|
1110032F04Rik
|
RIKEN cDNA 1110032F04 gene |
| chr10_+_128337761 | 0.49 |
ENSMUST00000005826.7
|
Cs
|
citrate synthase |
| chr12_+_84361968 | 0.49 |
ENSMUST00000021661.6
|
Coq6
|
coenzyme Q6 homolog (yeast) |
| chr2_-_45112890 | 0.48 |
ENSMUST00000076836.6
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr3_-_89349968 | 0.48 |
ENSMUST00000074582.6
ENSMUST00000107448.2 ENSMUST00000029676.5 |
Adam15
|
a disintegrin and metallopeptidase domain 15 (metargidin) |
| chr5_-_150518164 | 0.47 |
ENSMUST00000118769.1
|
Zar1l
|
zygote arrest 1-like |
| chr1_-_172027251 | 0.47 |
ENSMUST00000138714.1
|
Vangl2
|
vang-like 2 (van gogh, Drosophila) |
| chr7_-_19399859 | 0.46 |
ENSMUST00000047170.3
ENSMUST00000108459.2 |
Klc3
|
kinesin light chain 3 |
| chr5_+_92809372 | 0.46 |
ENSMUST00000113054.2
|
Shroom3
|
shroom family member 3 |
| chr7_-_30559600 | 0.46 |
ENSMUST00000043975.4
ENSMUST00000156241.1 |
Lin37
|
lin-37 homolog (C. elegans) |
| chr2_-_5676046 | 0.46 |
ENSMUST00000114987.3
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr3_-_89349939 | 0.44 |
ENSMUST00000107446.1
|
Adam15
|
a disintegrin and metallopeptidase domain 15 (metargidin) |
| chr7_-_30559828 | 0.43 |
ENSMUST00000108164.1
|
Lin37
|
lin-37 homolog (C. elegans) |
| chr4_-_149126688 | 0.43 |
ENSMUST00000030815.2
|
Cort
|
cortistatin |
| chr15_-_8710734 | 0.43 |
ENSMUST00000005493.7
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr5_-_66514815 | 0.43 |
ENSMUST00000161879.1
ENSMUST00000159357.1 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr5_-_34660068 | 0.43 |
ENSMUST00000041364.9
|
Nop14
|
NOP14 nucleolar protein |
| chr18_-_24603791 | 0.42 |
ENSMUST00000070726.3
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr12_+_51348019 | 0.42 |
ENSMUST00000054308.6
|
G2e3
|
G2/M-phase specific E3 ubiquitin ligase |
| chr7_+_25681158 | 0.42 |
ENSMUST00000108403.3
|
B9d2
|
B9 protein domain 2 |
| chr1_-_75046639 | 0.42 |
ENSMUST00000152855.1
|
Nhej1
|
nonhomologous end-joining factor 1 |
| chr16_+_22918378 | 0.42 |
ENSMUST00000170805.1
|
Fetub
|
fetuin beta |
| chr19_-_41933276 | 0.41 |
ENSMUST00000075280.4
ENSMUST00000112123.2 |
Exosc1
|
exosome component 1 |
| chr1_+_134405984 | 0.41 |
ENSMUST00000173908.1
|
Cyb5r1
|
cytochrome b5 reductase 1 |
| chr9_+_62858085 | 0.40 |
ENSMUST00000034777.6
ENSMUST00000163820.1 |
Calml4
|
calmodulin-like 4 |
| chr3_+_133338936 | 0.40 |
ENSMUST00000150386.1
ENSMUST00000125858.1 |
Ppa2
|
pyrophosphatase (inorganic) 2 |
| chr7_+_125829653 | 0.39 |
ENSMUST00000124223.1
|
D430042O09Rik
|
RIKEN cDNA D430042O09 gene |
| chr3_-_86548268 | 0.38 |
ENSMUST00000077524.3
|
Mab21l2
|
mab-21-like 2 (C. elegans) |
| chr7_-_46667303 | 0.37 |
ENSMUST00000168335.1
|
Tph1
|
tryptophan hydroxylase 1 |
| chr12_+_51348265 | 0.36 |
ENSMUST00000119211.1
|
G2e3
|
G2/M-phase specific E3 ubiquitin ligase |
| chr5_-_114773488 | 0.36 |
ENSMUST00000178440.1
ENSMUST00000043283.7 ENSMUST00000112185.2 |
Git2
|
G protein-coupled receptor kinase-interactor 2 |
| chr5_+_138187485 | 0.36 |
ENSMUST00000110934.2
|
Cnpy4
|
canopy 4 homolog (zebrafish) |
| chr9_-_106096776 | 0.36 |
ENSMUST00000121963.1
|
Col6a4
|
collagen, type VI, alpha 4 |
| chr1_-_55226768 | 0.36 |
ENSMUST00000027121.8
ENSMUST00000114428.2 |
Rftn2
|
raftlin family member 2 |
| chr6_+_124304646 | 0.35 |
ENSMUST00000112541.2
ENSMUST00000032234.2 |
Cd163
|
CD163 antigen |
| chr4_+_140701466 | 0.35 |
ENSMUST00000038893.5
ENSMUST00000138808.1 |
Rcc2
|
regulator of chromosome condensation 2 |
| chr7_-_101837776 | 0.35 |
ENSMUST00000165052.1
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr1_-_134955847 | 0.35 |
ENSMUST00000168381.1
|
Ppp1r12b
|
protein phosphatase 1, regulatory (inhibitor) subunit 12B |
| chr6_-_145865302 | 0.34 |
ENSMUST00000111703.1
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chrX_+_153126897 | 0.34 |
ENSMUST00000163801.1
|
Foxr2
|
forkhead box R2 |
| chr13_+_109685994 | 0.34 |
ENSMUST00000074103.5
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr8_-_36953139 | 0.34 |
ENSMUST00000179501.1
|
Dlc1
|
deleted in liver cancer 1 |
| chr12_+_3954943 | 0.33 |
ENSMUST00000020990.5
|
Pomc
|
pro-opiomelanocortin-alpha |
| chr2_+_125068118 | 0.32 |
ENSMUST00000070353.3
|
Slc24a5
|
solute carrier family 24, member 5 |
| chr3_+_18054258 | 0.32 |
ENSMUST00000026120.6
|
Bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr12_+_30884321 | 0.32 |
ENSMUST00000067087.6
|
Fam150b
|
family with sequence similarity 150, member B |
| chr3_-_50443603 | 0.32 |
ENSMUST00000029297.4
|
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr14_-_48665098 | 0.32 |
ENSMUST00000118578.1
|
Otx2
|
orthodenticle homolog 2 (Drosophila) |
| chrX_-_157415286 | 0.32 |
ENSMUST00000079945.4
ENSMUST00000138396.1 |
Phex
|
phosphate regulating gene with homologies to endopeptidases on the X chromosome (hypophosphatemia, vitamin D resistant rickets) |
| chr13_-_97747373 | 0.32 |
ENSMUST00000123535.1
|
5330416C01Rik
|
RIKEN cDNA 5330416C01 gene |
| chrM_+_14138 | 0.31 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr17_+_71019548 | 0.31 |
ENSMUST00000073211.5
ENSMUST00000179759.1 |
Myom1
|
myomesin 1 |
| chr2_+_3114220 | 0.31 |
ENSMUST00000072955.5
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr10_+_88146992 | 0.30 |
ENSMUST00000052355.7
|
Nup37
|
nucleoporin 37 |
| chr7_-_73541738 | 0.30 |
ENSMUST00000169922.2
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr2_+_69897220 | 0.29 |
ENSMUST00000055758.9
ENSMUST00000112251.2 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr15_-_66812593 | 0.29 |
ENSMUST00000100572.3
|
Sla
|
src-like adaptor |
| chr2_-_85196697 | 0.29 |
ENSMUST00000099930.2
ENSMUST00000111601.1 |
Lrrc55
|
leucine rich repeat containing 55 |
| chr2_+_69897255 | 0.28 |
ENSMUST00000131553.1
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr17_+_71019503 | 0.28 |
ENSMUST00000024847.7
|
Myom1
|
myomesin 1 |
| chr12_+_51348370 | 0.28 |
ENSMUST00000121521.1
|
G2e3
|
G2/M-phase specific E3 ubiquitin ligase |
| chr4_-_96785186 | 0.28 |
ENSMUST00000107071.1
|
Gm12695
|
predicted gene 12695 |
| chrX_-_150589844 | 0.28 |
ENSMUST00000112725.1
ENSMUST00000112720.1 |
Apex2
|
apurinic/apyrimidinic endonuclease 2 |
| chr17_-_70853482 | 0.28 |
ENSMUST00000118283.1
|
Tgif1
|
TGFB-induced factor homeobox 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mnx1_Lhx6_Lmx1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.9 | 2.6 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.8 | 4.2 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.8 | 2.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.6 | 1.8 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.4 | 2.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.4 | 1.2 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.4 | 1.2 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.3 | 1.0 | GO:2000852 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.3 | 0.9 | GO:0061743 | motor learning(GO:0061743) |
| 0.3 | 1.2 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.3 | 3.9 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.3 | 1.8 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.3 | 1.4 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.3 | 1.5 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.3 | 0.8 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.3 | 5.8 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 1.7 | GO:0042747 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) |
| 0.2 | 0.9 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.2 | 0.9 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.2 | 0.7 | GO:0061055 | myotome development(GO:0061055) |
| 0.2 | 0.6 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.2 | 0.7 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.2 | 1.2 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.2 | 4.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 1.6 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.9 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.1 | 0.6 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.3 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.1 | 0.3 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.1 | 0.8 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 1.0 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 | 0.6 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) |
| 0.1 | 2.1 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 1.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 1.7 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.5 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 | 0.8 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.6 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.1 | 1.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 1.9 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 1.0 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 | 0.3 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.3 | GO:1904245 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) |
| 0.1 | 0.9 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.1 | 1.0 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.1 | 1.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.3 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) |
| 0.1 | 0.5 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.3 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 1.0 | GO:0061377 | mammary gland alveolus development(GO:0060749) mammary gland lobule development(GO:0061377) |
| 0.1 | 1.2 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 0.2 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.1 | 0.8 | GO:0098596 | vocal learning(GO:0042297) imitative learning(GO:0098596) |
| 0.1 | 0.9 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.1 | 1.4 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 0.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.2 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.1 | 1.8 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 0.3 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.1 | 2.8 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 1.0 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 0.2 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.3 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 1.4 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.1 | 0.4 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.1 | 0.5 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:1900239 | regulation of phenotypic switching(GO:1900239) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.0 | 1.5 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 0.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.5 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.3 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.6 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.7 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 1.8 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.1 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.3 | GO:0021957 | corpus callosum morphogenesis(GO:0021540) corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 0.5 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.2 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.3 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.3 | GO:0042635 | positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 1.5 | GO:0007595 | lactation(GO:0007595) |
| 0.0 | 0.7 | GO:1901016 | regulation of potassium ion transmembrane transporter activity(GO:1901016) |
| 0.0 | 3.3 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 0.2 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 1.0 | GO:0061050 | regulation of cell growth involved in cardiac muscle cell development(GO:0061050) |
| 0.0 | 0.4 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.1 | GO:1905065 | cysteine biosynthetic process via cystathionine(GO:0019343) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.0 | 1.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.2 | GO:0046601 | positive regulation of centriole replication(GO:0046601) de novo centriole assembly(GO:0098535) |
| 0.0 | 0.9 | GO:0071385 | cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.4 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.9 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.0 | 0.1 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.1 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.5 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 1.0 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.7 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.2 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.3 | GO:0021756 | striatum development(GO:0021756) |
| 0.0 | 0.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.4 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 1.8 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 0.4 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.0 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.3 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.3 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.2 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 3.1 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.2 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.4 | GO:0006953 | acute-phase response(GO:0006953) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.4 | GO:0031523 | Myb complex(GO:0031523) |
| 0.6 | 4.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.6 | 5.8 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.4 | 2.7 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.3 | 1.2 | GO:0008623 | CHRAC(GO:0008623) |
| 0.3 | 1.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.3 | 5.8 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.2 | 1.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 1.0 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.1 | 0.5 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 0.7 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 1.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 2.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.4 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 1.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.8 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.4 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 3.2 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 0.8 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.6 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 1.0 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.0 | 1.0 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.3 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 3.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.5 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.5 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 1.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 3.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 2.4 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 1.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.5 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.9 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 2.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 3.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.0 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 1.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.7 | 3.4 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.4 | 1.2 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.3 | 1.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 1.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.2 | 2.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.2 | 1.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 0.7 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.2 | 0.9 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 2.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 1.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 1.0 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 3.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 3.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.8 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.3 | GO:0071820 | N-box binding(GO:0071820) |
| 0.1 | 0.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 2.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.3 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.1 | 0.6 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.3 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 2.9 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 0.3 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.1 | 0.5 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.7 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 2.0 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 2.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.8 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 3.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.5 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 0.9 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 1.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.3 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 2.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.3 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 2.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.7 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.6 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.5 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.3 | GO:0046030 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 0.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.1 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 1.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.1 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.0 | 0.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 1.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 3.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 4.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 8.0 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 1.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 1.3 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 4.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 1.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 1.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 2.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 3.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 2.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 0.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 2.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 7.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 2.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 0.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.9 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 1.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.7 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |