Project
avrg: 12D miR HR13_24
Navigation
Downloads
Results for Nr0b1
Z-value: 0.49
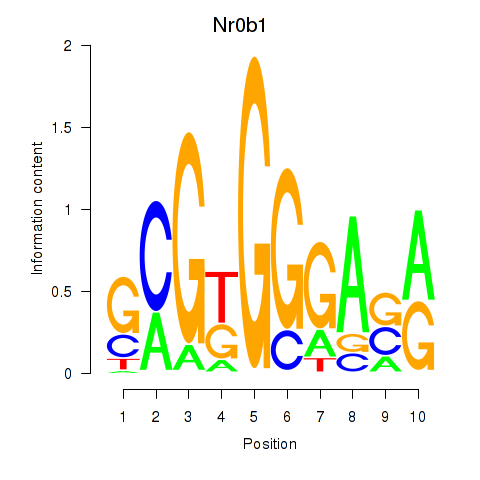
Transcription factors associated with Nr0b1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr0b1
|
ENSMUSG00000025056.4 | nuclear receptor subfamily 0, group B, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr0b1 | mm10_v2_chrX_+_86191764_86191782 | -0.59 | 5.5e-02 | Click! |
Activity profile of Nr0b1 motif
Sorted Z-values of Nr0b1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_144332146 | 0.76 |
ENSMUST00000037423.3
|
Ovol2
|
ovo-like 2 (Drosophila) |
| chr2_-_144331695 | 0.70 |
ENSMUST00000103171.3
|
Ovol2
|
ovo-like 2 (Drosophila) |
| chr4_-_11386679 | 0.68 |
ENSMUST00000043781.7
ENSMUST00000108310.1 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr5_+_30913398 | 0.41 |
ENSMUST00000031055.5
|
Emilin1
|
elastin microfibril interfacer 1 |
| chr4_-_11386394 | 0.39 |
ENSMUST00000155519.1
|
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr7_+_19359740 | 0.37 |
ENSMUST00000140836.1
|
Ppp1r13l
|
protein phosphatase 1, regulatory (inhibitor) subunit 13 like |
| chr2_-_65238573 | 0.30 |
ENSMUST00000090896.3
ENSMUST00000155082.1 |
Cobll1
|
Cobl-like 1 |
| chr2_-_65238625 | 0.30 |
ENSMUST00000112429.2
ENSMUST00000102726.1 ENSMUST00000112430.1 |
Cobll1
|
Cobl-like 1 |
| chr4_+_116221590 | 0.27 |
ENSMUST00000147292.1
|
Pik3r3
|
phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 3 (p55) |
| chr15_+_98634743 | 0.26 |
ENSMUST00000003442.7
|
Cacnb3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr4_+_116221689 | 0.25 |
ENSMUST00000106490.2
|
Pik3r3
|
phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 3 (p55) |
| chr7_+_19094594 | 0.24 |
ENSMUST00000049454.5
|
Six5
|
sine oculis-related homeobox 5 |
| chr4_+_116221633 | 0.24 |
ENSMUST00000030464.7
|
Pik3r3
|
phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 3 (p55) |
| chr5_-_107726017 | 0.21 |
ENSMUST00000159263.2
|
Gfi1
|
growth factor independent 1 |
| chr16_-_20621255 | 0.21 |
ENSMUST00000052939.2
|
Camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr2_+_71786923 | 0.20 |
ENSMUST00000112101.1
ENSMUST00000028522.3 |
Itga6
|
integrin alpha 6 |
| chr9_+_80165079 | 0.19 |
ENSMUST00000184480.1
|
Myo6
|
myosin VI |
| chr2_+_157279065 | 0.18 |
ENSMUST00000029171.5
|
Rpn2
|
ribophorin II |
| chr16_-_45844303 | 0.18 |
ENSMUST00000036355.6
|
Phldb2
|
pleckstrin homology-like domain, family B, member 2 |
| chr16_-_45844228 | 0.18 |
ENSMUST00000076333.5
|
Phldb2
|
pleckstrin homology-like domain, family B, member 2 |
| chr6_+_134830145 | 0.17 |
ENSMUST00000046303.5
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr3_-_152166230 | 0.16 |
ENSMUST00000046614.9
|
Gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr17_+_29490812 | 0.16 |
ENSMUST00000024811.6
|
Pim1
|
proviral integration site 1 |
| chrX_-_145505175 | 0.15 |
ENSMUST00000143610.1
|
Amot
|
angiomotin |
| chr1_+_88087802 | 0.15 |
ENSMUST00000113139.1
|
Ugt1a8
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr6_+_28981490 | 0.14 |
ENSMUST00000164104.1
|
Gm3294
|
predicted gene 3294 |
| chr6_+_134830216 | 0.14 |
ENSMUST00000111937.1
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr2_+_157279026 | 0.13 |
ENSMUST00000116380.2
|
Rpn2
|
ribophorin II |
| chr9_+_80165013 | 0.13 |
ENSMUST00000035889.8
ENSMUST00000113268.1 |
Myo6
|
myosin VI |
| chr5_+_64803513 | 0.11 |
ENSMUST00000165536.1
|
Klf3
|
Kruppel-like factor 3 (basic) |
| chr2_+_129198757 | 0.11 |
ENSMUST00000028880.3
|
Slc20a1
|
solute carrier family 20, member 1 |
| chr17_-_80728026 | 0.10 |
ENSMUST00000112389.2
ENSMUST00000025089.7 |
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr11_+_94328242 | 0.09 |
ENSMUST00000021227.5
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr15_+_35371498 | 0.09 |
ENSMUST00000048646.7
|
Vps13b
|
vacuolar protein sorting 13B (yeast) |
| chr16_+_44173239 | 0.09 |
ENSMUST00000119746.1
|
Gm608
|
predicted gene 608 |
| chr13_-_64274879 | 0.08 |
ENSMUST00000109770.1
|
Cdc14b
|
CDC14 cell division cycle 14B |
| chr13_-_31559333 | 0.08 |
ENSMUST00000170573.1
|
A530084C06Rik
|
RIKEN cDNA A530084C06 gene |
| chr3_+_86224665 | 0.07 |
ENSMUST00000107635.1
|
Lrba
|
LPS-responsive beige-like anchor |
| chr13_-_94358818 | 0.07 |
ENSMUST00000059598.2
|
Gm9776
|
predicted gene 9776 |
| chr8_-_95434869 | 0.07 |
ENSMUST00000034249.6
|
Gtl3
|
gene trap locus 3 |
| chr6_+_47454320 | 0.07 |
ENSMUST00000031697.8
|
Cul1
|
cullin 1 |
| chr3_+_89715016 | 0.07 |
ENSMUST00000098924.2
|
Adar
|
adenosine deaminase, RNA-specific |
| chr2_+_153842981 | 0.06 |
ENSMUST00000175856.1
|
Efcab8
|
EF-hand calcium binding domain 8 |
| chr5_+_31193227 | 0.06 |
ENSMUST00000031029.8
ENSMUST00000133711.1 ENSMUST00000132471.1 |
Snx17
|
sorting nexin 17 |
| chr11_+_107547925 | 0.06 |
ENSMUST00000100305.1
ENSMUST00000075012.1 ENSMUST00000106746.1 |
Helz
|
helicase with zinc finger domain |
| chr10_+_39732099 | 0.05 |
ENSMUST00000019986.6
|
Rev3l
|
REV3-like, catalytic subunit of DNA polymerase zeta RAD54 like (S. cerevisiae) |
| chr7_-_65371210 | 0.05 |
ENSMUST00000102592.3
|
Tjp1
|
tight junction protein 1 |
| chr4_-_129696817 | 0.05 |
ENSMUST00000102588.3
|
Tmem39b
|
transmembrane protein 39b |
| chr15_-_81697256 | 0.05 |
ENSMUST00000072910.5
|
Chadl
|
chondroadherin-like |
| chr3_-_107458895 | 0.05 |
ENSMUST00000009617.8
|
Kcnc4
|
potassium voltage gated channel, Shaw-related subfamily, member 4 |
| chr17_-_26508463 | 0.05 |
ENSMUST00000025025.6
|
Dusp1
|
dual specificity phosphatase 1 |
| chr17_+_88530113 | 0.05 |
ENSMUST00000038551.6
|
Ppp1r21
|
protein phosphatase 1, regulatory subunit 21 |
| chr2_-_80581234 | 0.05 |
ENSMUST00000028386.5
|
Nckap1
|
NCK-associated protein 1 |
| chr11_+_94327984 | 0.05 |
ENSMUST00000107818.2
ENSMUST00000051221.6 |
Ankrd40
|
ankyrin repeat domain 40 |
| chr3_+_54156039 | 0.04 |
ENSMUST00000029311.6
|
Trpc4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr10_-_81350389 | 0.04 |
ENSMUST00000020454.4
ENSMUST00000105324.2 ENSMUST00000154609.2 ENSMUST00000105323.1 |
Hmg20b
|
high mobility group 20B |
| chr18_+_35829798 | 0.04 |
ENSMUST00000060722.6
|
Cxxc5
|
CXXC finger 5 |
| chr10_-_128589650 | 0.04 |
ENSMUST00000082059.6
|
Erbb3
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) |
| chr6_-_52191695 | 0.04 |
ENSMUST00000101395.2
|
Hoxa4
|
homeobox A4 |
| chr9_+_110333402 | 0.04 |
ENSMUST00000133114.1
ENSMUST00000125759.1 |
Scap
|
SREBF chaperone |
| chr5_+_121711609 | 0.04 |
ENSMUST00000051950.7
|
Atxn2
|
ataxin 2 |
| chr13_-_14063395 | 0.03 |
ENSMUST00000170957.1
|
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr4_-_132422484 | 0.03 |
ENSMUST00000102568.3
|
Phactr4
|
phosphatase and actin regulator 4 |
| chr3_-_123690806 | 0.03 |
ENSMUST00000154668.1
|
Ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr2_-_157279519 | 0.03 |
ENSMUST00000143663.1
|
Mroh8
|
maestro heat-like repeat family member 8 |
| chr4_-_118134869 | 0.03 |
ENSMUST00000097912.1
ENSMUST00000030263.2 ENSMUST00000106410.1 |
St3gal3
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
| chr2_-_80581380 | 0.03 |
ENSMUST00000111760.2
|
Nckap1
|
NCK-associated protein 1 |
| chr7_+_79500081 | 0.03 |
ENSMUST00000181511.2
ENSMUST00000182937.1 |
AI854517
|
expressed sequence AI854517 |
| chr17_+_34898463 | 0.03 |
ENSMUST00000114033.2
ENSMUST00000078061.6 |
Ehmt2
|
euchromatic histone lysine N-methyltransferase 2 |
| chr4_-_132422394 | 0.02 |
ENSMUST00000152271.1
ENSMUST00000084170.5 |
Phactr4
|
phosphatase and actin regulator 4 |
| chr15_+_80711292 | 0.02 |
ENSMUST00000067689.7
|
Tnrc6b
|
trinucleotide repeat containing 6b |
| chr13_+_94358943 | 0.02 |
ENSMUST00000022196.3
|
Ap3b1
|
adaptor-related protein complex 3, beta 1 subunit |
| chr15_-_38078842 | 0.02 |
ENSMUST00000110336.2
|
Ubr5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr8_+_71371283 | 0.02 |
ENSMUST00000110051.1
ENSMUST00000002469.8 ENSMUST00000110052.1 |
Ocel1
|
occludin/ELL domain containing 1 |
| chr1_+_162570687 | 0.01 |
ENSMUST00000050010.4
ENSMUST00000150040.1 |
Vamp4
|
vesicle-associated membrane protein 4 |
| chr4_-_123750236 | 0.01 |
ENSMUST00000102636.3
|
Akirin1
|
akirin 1 |
| chr11_+_105181527 | 0.01 |
ENSMUST00000106941.2
|
Tlk2
|
tousled-like kinase 2 (Arabidopsis) |
| chr10_+_90829538 | 0.01 |
ENSMUST00000179694.2
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr1_+_140246216 | 0.01 |
ENSMUST00000119786.1
ENSMUST00000120796.1 ENSMUST00000060201.8 ENSMUST00000120709.1 |
Kcnt2
|
potassium channel, subfamily T, member 2 |
| chr5_-_137531952 | 0.01 |
ENSMUST00000140139.1
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr9_-_44881274 | 0.01 |
ENSMUST00000002095.3
ENSMUST00000114689.1 ENSMUST00000128768.1 |
Kmt2a
|
lysine (K)-specific methyltransferase 2A |
| chr2_+_59160884 | 0.01 |
ENSMUST00000037903.8
|
Pkp4
|
plakophilin 4 |
| chr19_+_45015198 | 0.00 |
ENSMUST00000179108.1
|
Lzts2
|
leucine zipper, putative tumor suppressor 2 |
| chr14_+_70457447 | 0.00 |
ENSMUST00000003561.3
|
Phyhip
|
phytanoyl-CoA hydroxylase interacting protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr0b1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.1 | 0.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0070103 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.0 | 0.1 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 1.1 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.3 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.2 | GO:2000138 | vitamin D receptor signaling pathway(GO:0070561) positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 | 0.3 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.0 | 0.2 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.0 | 0.0 | GO:1904057 | negative regulation of sensory perception of pain(GO:1904057) |
| 0.0 | 0.8 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.4 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.1 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.1 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.0 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 0.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.3 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.0 | 0.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |