Project
DANIO-CODE
Navigation
Downloads
Results for CABZ01093502.2
Z-value: 0.21
Transcription factors associated with CABZ01093502.2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
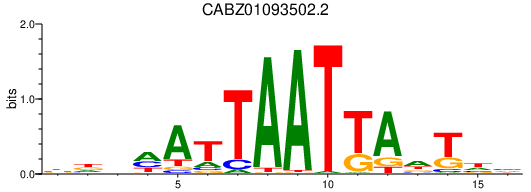
CABZ01093502.2
|
ENSDARG00000105053 | developing brain homeobox 2 |
Activity profile of CABZ01093502.2 motif
Sorted Z-values of CABZ01093502.2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CABZ01093502.2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_21982644 | 0.38 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr9_+_21982679 | 0.35 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr11_+_17849608 | 0.30 |
ENSDART00000020283
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr2_+_30800532 | 0.26 |
|
|
|
| chr9_+_21982756 | 0.26 |
ENSDART00000059652
|
rev1
|
REV1, polymerase (DNA directed) |
| chr3_-_39912816 | 0.23 |
ENSDART00000102540
|
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr22_+_2735606 | 0.23 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr15_-_9443212 | 0.23 |
ENSDART00000177158
|
sacs
|
sacsin molecular chaperone |
| chr7_-_26226924 | 0.22 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr5_-_3673698 | 0.22 |
|
|
|
| chr4_-_14981572 | 0.18 |
|
|
|
| chr21_-_33998091 | 0.16 |
ENSDART00000047515
ENSDART00000138575 ENSDART00000170749 |
rnf145b
|
ring finger protein 145b |
| chr21_-_13026036 | 0.16 |
ENSDART00000135623
|
fam219aa
|
family with sequence similarity 219, member Aa |
| chr8_-_20198473 | 0.15 |
ENSDART00000063400
|
mllt1a
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 1a |
| chr7_-_26226628 | 0.15 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr17_-_42523585 | 0.15 |
ENSDART00000154755
|
prkd3
|
protein kinase D3 |
| chr8_-_39850553 | 0.13 |
ENSDART00000131372
|
mlec
|
malectin |
| chr21_+_5821072 | 0.13 |
ENSDART00000121769
|
rexo4
|
REX4 homolog, 3'-5' exonuclease |
| chr19_+_41894863 | 0.13 |
ENSDART00000087187
|
ago2
|
argonaute RISC catalytic component 2 |
| chr15_-_43402935 | 0.12 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr19_+_1743359 | 0.12 |
ENSDART00000166744
|
dennd3a
|
DENN/MADD domain containing 3a |
| chr21_+_43177310 | 0.12 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr21_-_13026077 | 0.12 |
ENSDART00000024616
|
fam219aa
|
family with sequence similarity 219, member Aa |
| chr5_+_12894381 | 0.11 |
ENSDART00000051669
|
tctn2
|
tectonic family member 2 |
| chr9_+_24255311 | 0.11 |
ENSDART00000166303
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr20_+_40560443 | 0.11 |
ENSDART00000153119
|
serinc1
|
serine incorporator 1 |
| chr21_-_33998117 | 0.11 |
ENSDART00000047515
ENSDART00000138575 ENSDART00000170749 |
rnf145b
|
ring finger protein 145b |
| chr2_+_11094785 | 0.11 |
ENSDART00000091497
|
tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr2_-_11094083 | 0.11 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr25_-_20933145 | 0.11 |
ENSDART00000138985
ENSDART00000046298 |
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr6_+_55250892 | 0.09 |
|
|
|
| chr13_+_35402766 | 0.09 |
ENSDART00000163368
ENSDART00000075414 ENSDART00000112947 |
wdr27
|
WD repeat domain 27 |
| chr8_+_2471513 | 0.09 |
ENSDART00000063943
|
mrpl40
|
mitochondrial ribosomal protein L40 |
| chr7_-_57206977 | 0.09 |
ENSDART00000147036
|
sirt3
|
sirtuin 3 |
| chr1_-_358810 | 0.09 |
ENSDART00000098627
|
pros1
|
protein S (alpha) |
| chr1_+_51087450 | 0.08 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr20_+_25687135 | 0.07 |
ENSDART00000063122
|
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr9_+_24255064 | 0.07 |
ENSDART00000101577
ENSDART00000159324 ENSDART00000023196 ENSDART00000079689 ENSDART00000172743 ENSDART00000171577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr24_-_14447519 | 0.07 |
|
|
|
| chr3_+_25036636 | 0.07 |
ENSDART00000077493
|
TST
|
thiosulfate sulfurtransferase |
| chr8_+_50964745 | 0.06 |
ENSDART00000013870
|
ENSDARG00000007359
|
ENSDARG00000007359 |
| chr8_-_2471264 | 0.06 |
ENSDART00000056767
|
acaa2
|
acetyl-CoA acyltransferase 2 |
| chr16_+_51597409 | 0.06 |
|
|
|
| chr24_-_14447655 | 0.05 |
|
|
|
| chr21_-_33998051 | 0.05 |
ENSDART00000047515
ENSDART00000138575 ENSDART00000170749 |
rnf145b
|
ring finger protein 145b |
| chr4_+_75469488 | 0.04 |
ENSDART00000144892
|
znf1009
|
zinc finger protein 1009 |
| chr14_+_37317006 | 0.03 |
|
|
|
| chr17_-_12610211 | 0.03 |
ENSDART00000003418
ENSDART00000168390 |
brms1la
|
breast cancer metastasis-suppressor 1-like a |
| chr4_-_2370333 | 0.02 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr24_-_14447136 | 0.02 |
|
|
|
| chr11_+_17849528 | 0.01 |
ENSDART00000020283
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr3_+_25036265 | 0.01 |
ENSDART00000077493
|
TST
|
thiosulfate sulfurtransferase |
| chr19_+_46567459 | 0.01 |
ENSDART00000164055
|
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr13_+_35402382 | 0.01 |
ENSDART00000163368
|
wdr27
|
WD repeat domain 27 |
| chr9_-_51976213 | 0.00 |
ENSDART00000006612
|
tbr1b
|
T-box, brain, 1b |
| chr11_+_11284030 | 0.00 |
ENSDART00000026814
|
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.2 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.3 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.0 | 0.1 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:1990748 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0017125 | deoxycytidyl transferase activity(GO:0017125) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0070551 | endoribonuclease activity, cleaving siRNA-paired mRNA(GO:0070551) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |