Project
DANIO-CODE
Navigation
Downloads
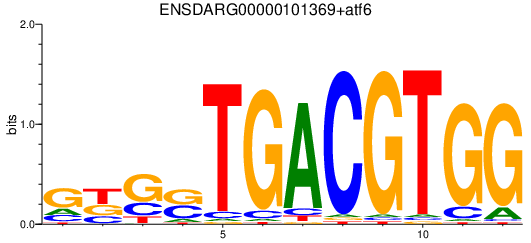
Results for ENSDARG00000101369+atf6
Z-value: 1.17
Transcription factors associated with ENSDARG00000101369+atf6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
atf6
|
ENSDARG00000012656 | activating transcription factor 6 |
|
ENSDARG00000101369
|
ENSDARG00000101369 | ENSDARG00000101369 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| atf6 | dr10_dc_chr20_-_33961862_33961999 | -0.47 | 6.3e-02 | Click! |
Activity profile of ENSDARG00000101369+atf6 motif
Sorted Z-values of ENSDARG00000101369+atf6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ENSDARG00000101369+atf6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_9036049 | 2.33 |
ENSDART00000160079
|
gak
|
cyclin G associated kinase |
| chr3_-_31713951 | 1.53 |
ENSDART00000157028
|
map3k3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr10_+_15644868 | 1.42 |
ENSDART00000139259
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr25_-_36512943 | 1.38 |
ENSDART00000114508
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr11_-_22663430 | 1.38 |
ENSDART00000154813
|
mdm4
|
MDM4, p53 regulator |
| chr3_-_31713730 | 1.29 |
ENSDART00000157028
|
map3k3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr25_-_17282381 | 1.23 |
ENSDART00000178173
|
cyp2x7
|
cytochrome P450, family 2, subfamily X, polypeptide 7 |
| chr15_-_43954665 | 1.20 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr7_-_73613531 | 1.18 |
ENSDART00000128137
|
zgc:92594
|
zgc:92594 |
| chr7_-_73613763 | 1.17 |
ENSDART00000128137
|
zgc:92594
|
zgc:92594 |
| chr18_+_39506453 | 1.15 |
ENSDART00000126978
|
acadl
|
acyl-CoA dehydrogenase, long chain |
| chr21_+_10609580 | 1.13 |
ENSDART00000102304
|
lman1
|
lectin, mannose-binding, 1 |
| chr17_+_21082384 | 1.13 |
ENSDART00000035432
|
entpd6
|
ectonucleoside triphosphate diphosphohydrolase 6 (putative) |
| chr16_+_2327771 | 1.10 |
ENSDART00000171378
|
preb
|
prolactin regulatory element binding |
| chr22_+_2014149 | 1.09 |
ENSDART00000165168
|
znf1157
|
zinc finger protein 1157 |
| chr23_-_25871518 | 1.08 |
ENSDART00000041833
|
fitm2
|
fat storage-inducing transmembrane protein 2 |
| chr17_+_21082416 | 1.08 |
ENSDART00000035432
|
entpd6
|
ectonucleoside triphosphate diphosphohydrolase 6 (putative) |
| chr17_-_24684870 | 1.07 |
ENSDART00000156061
|
si:ch211-15d5.12
|
si:ch211-15d5.12 |
| chr5_-_9035835 | 1.05 |
ENSDART00000160079
|
gak
|
cyclin G associated kinase |
| chr22_+_1445302 | 1.04 |
ENSDART00000164685
|
si:dkeyp-53d3.5
|
si:dkeyp-53d3.5 |
| chr3_-_26052785 | 1.03 |
ENSDART00000147517
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr7_+_34923319 | 1.02 |
|
|
|
| chr9_+_8920413 | 1.01 |
ENSDART00000147820
|
carkd
|
carbohydrate kinase domain containing |
| chr4_-_27109874 | 1.00 |
ENSDART00000146459
|
creld2
|
cysteine-rich with EGF-like domains 2 |
| chr11_-_24300905 | 0.99 |
ENSDART00000171004
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr7_+_13238684 | 0.97 |
ENSDART00000053535
|
arih1l
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 like |
| chr22_+_2735606 | 0.97 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr18_+_8959686 | 0.92 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr7_-_52850565 | 0.92 |
|
|
|
| chr11_-_22663375 | 0.90 |
ENSDART00000154813
|
mdm4
|
MDM4, p53 regulator |
| chr5_-_41204170 | 0.90 |
ENSDART00000134492
|
alkbh2
|
alkB homolog 2, alpha-ketoglutarate-dependent dioxygenase |
| chr6_+_49902765 | 0.89 |
ENSDART00000023515
|
chmp4ba
|
charged multivesicular body protein 4Ba |
| chr9_+_21982644 | 0.88 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr24_-_18540597 | 0.87 |
ENSDART00000176165
|
zgc:66014
|
zgc:66014 |
| chr6_-_11576632 | 0.86 |
ENSDART00000151717
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr9_+_24381412 | 0.86 |
ENSDART00000003482
|
stk17b
|
serine/threonine kinase 17b (apoptosis-inducing) |
| chr10_+_2554651 | 0.85 |
ENSDART00000016103
|
nxnl2
|
nucleoredoxin-like 2 |
| chr20_+_46524223 | 0.84 |
ENSDART00000153290
|
rad51
|
RAD51 recombinase |
| chr3_+_59972297 | 0.84 |
|
|
|
| chr22_+_1837102 | 0.83 |
ENSDART00000163288
|
znf1174
|
zinc finger protein 1174 |
| chr7_+_69211965 | 0.82 |
ENSDART00000028064
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr9_+_24381641 | 0.82 |
ENSDART00000003482
|
stk17b
|
serine/threonine kinase 17b (apoptosis-inducing) |
| chr10_-_8839096 | 0.82 |
ENSDART00000080763
|
si:dkey-27b3.2
|
si:dkey-27b3.2 |
| chr9_+_21982679 | 0.81 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr23_+_20505072 | 0.81 |
ENSDART00000132920
|
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr3_+_23964015 | 0.80 |
ENSDART00000138270
|
copz2
|
coatomer protein complex, subunit zeta 2 |
| chr12_+_33818414 | 0.79 |
ENSDART00000085888
|
trim8b
|
tripartite motif containing 8b |
| chr22_+_1445227 | 0.79 |
ENSDART00000164685
|
si:dkeyp-53d3.5
|
si:dkeyp-53d3.5 |
| chr8_+_5222023 | 0.79 |
ENSDART00000035676
|
bnip3la
|
BCL2/adenovirus E1B interacting protein 3-like a |
| chr9_-_27585644 | 0.79 |
ENSDART00000101401
|
tex30
|
testis expressed 30 |
| chr25_-_24150388 | 0.79 |
ENSDART00000073527
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr6_-_28952426 | 0.78 |
ENSDART00000126524
|
tbc1d23
|
TBC1 domain family, member 23 |
| chr14_-_46608307 | 0.78 |
ENSDART00000074038
|
scyl1
|
SCY1-like, kinase-like 1 |
| chr5_-_9035789 | 0.77 |
ENSDART00000124384
|
gak
|
cyclin G associated kinase |
| chr25_+_21001126 | 0.77 |
ENSDART00000079012
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr2_+_10265922 | 0.73 |
ENSDART00000143876
|
cmpk
|
cytidylate kinase |
| chr17_+_17745119 | 0.71 |
ENSDART00000138189
ENSDART00000105013 |
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr18_+_8362265 | 0.71 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr3_-_26052601 | 0.71 |
ENSDART00000147517
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr23_+_20909885 | 0.70 |
ENSDART00000127628
|
errfi1b
|
ERBB receptor feedback inhibitor 1b |
| chr17_-_24860499 | 0.70 |
ENSDART00000123147
|
zbtb8a
|
zinc finger and BTB domain containing 8A |
| chr18_-_26131127 | 0.68 |
ENSDART00000163226
ENSDART00000004692 |
idh2
|
isocitrate dehydrogenase 2 (NADP+), mitochondrial |
| chr11_-_22663526 | 0.66 |
ENSDART00000155046
|
mdm4
|
MDM4, p53 regulator |
| chr25_-_36513113 | 0.66 |
ENSDART00000025494
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr25_-_24150237 | 0.65 |
ENSDART00000073527
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr21_+_10609341 | 0.64 |
ENSDART00000102304
|
lman1
|
lectin, mannose-binding, 1 |
| chr8_+_23840165 | 0.64 |
ENSDART00000037109
|
srpk1a
|
SRSF protein kinase 1a |
| chr23_-_31339986 | 0.63 |
ENSDART00000139746
|
si:dkey-261l7.2
|
si:dkey-261l7.2 |
| chr11_+_31362089 | 0.63 |
ENSDART00000158494
ENSDART00000173083 |
FO704610.1
|
ENSDARG00000100400 |
| chr5_-_29916352 | 0.63 |
ENSDART00000014666
|
arcn1a
|
archain 1a |
| chr10_-_28494204 | 0.62 |
ENSDART00000131220
|
btg3
|
B-cell translocation gene 3 |
| chr5_-_27394386 | 0.59 |
ENSDART00000171611
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr9_-_137883 | 0.59 |
ENSDART00000169377
|
FQ377903.3
|
ENSDARG00000104933 |
| chr22_+_32274241 | 0.59 |
ENSDART00000092082
|
manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr20_+_27041190 | 0.59 |
ENSDART00000164027
|
cdca4
|
cell division cycle associated 4 |
| chr15_+_17164535 | 0.58 |
ENSDART00000155350
|
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr23_+_20505319 | 0.58 |
ENSDART00000140219
|
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr10_+_16078021 | 0.58 |
ENSDART00000065037
ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr11_-_24300849 | 0.57 |
ENSDART00000171004
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr18_+_8362131 | 0.56 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr25_-_17282337 | 0.56 |
ENSDART00000064586
|
cyp2x7
|
cytochrome P450, family 2, subfamily X, polypeptide 7 |
| chr1_+_25911314 | 0.56 |
ENSDART00000011645
|
coro2a
|
coronin, actin binding protein, 2A |
| chr5_-_1432791 | 0.56 |
ENSDART00000128274
|
golga2
|
golgin A2 |
| chr9_+_21982756 | 0.56 |
ENSDART00000059652
|
rev1
|
REV1, polymerase (DNA directed) |
| chr12_+_31668126 | 0.55 |
ENSDART00000105584
|
lrrc59
|
leucine rich repeat containing 59 |
| chr17_+_25163592 | 0.55 |
ENSDART00000100277
|
cln8
|
CLN8, transmembrane ER and ERGIC protein |
| chr7_+_69212246 | 0.54 |
ENSDART00000159799
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr8_-_12631385 | 0.54 |
ENSDART00000143155
|
CU639468.1
|
ENSDARG00000093851 |
| chr1_+_36039889 | 0.54 |
ENSDART00000149022
|
arhgap10
|
Rho GTPase activating protein 10 |
| chr5_-_56254353 | 0.54 |
ENSDART00000014028
|
ppm1db
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Db |
| chr15_-_43954596 | 0.53 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr20_+_54545050 | 0.53 |
|
|
|
| chr15_-_31296524 | 0.52 |
ENSDART00000008854
|
wsb1
|
WD repeat and SOCS box containing 1 |
| chr10_+_16078433 | 0.52 |
ENSDART00000065037
ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr4_-_25526284 | 0.52 |
ENSDART00000142276
|
rbm17
|
RNA binding motif protein 17 |
| chr23_+_33792145 | 0.51 |
ENSDART00000024695
|
dazap2
|
DAZ associated protein 2 |
| chr11_+_413753 | 0.51 |
ENSDART00000082517
|
rab43
|
RAB43, member RAS oncogene family |
| chr3_+_34051311 | 0.51 |
ENSDART00000055252
|
TIMM29
|
translocase of inner mitochondrial membrane 29 |
| chr3_-_48458042 | 0.51 |
ENSDART00000156822
|
cdip1
|
cell death-inducing p53 target 1 |
| chr22_+_1451958 | 0.51 |
|
|
|
| chr5_-_48028931 | 0.50 |
ENSDART00000031194
|
lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr25_+_21001016 | 0.50 |
ENSDART00000079012
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr16_-_46600314 | 0.50 |
ENSDART00000160790
|
si:dkey-152b24.7
|
si:dkey-152b24.7 |
| chr17_+_9934845 | 0.50 |
ENSDART00000168055
|
sec23a
|
Sec23 homolog A, COPII coat complex component |
| chr22_+_2785213 | 0.49 |
ENSDART00000132419
|
si:dkey-20i20.7
|
si:dkey-20i20.7 |
| chr22_+_2386743 | 0.48 |
ENSDART00000132925
|
zgc:112977
|
zgc:112977 |
| chr7_+_69211741 | 0.48 |
ENSDART00000109644
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr6_-_50686517 | 0.48 |
ENSDART00000134146
|
mtss1
|
metastasis suppressor 1 |
| chr18_+_20493237 | 0.46 |
ENSDART00000128139
|
kbtbd4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr18_+_5066229 | 0.45 |
ENSDART00000157671
|
CABZ01080599.1
|
ENSDARG00000100626 |
| chr25_+_15983733 | 0.45 |
ENSDART00000165598
ENSDART00000061753 |
far1
|
fatty acyl CoA reductase 1 |
| chr20_-_50211708 | 0.45 |
ENSDART00000148892
|
extl3
|
exostosin-like glycosyltransferase 3 |
| chr12_+_31668168 | 0.44 |
ENSDART00000105584
|
lrrc59
|
leucine rich repeat containing 59 |
| chr18_-_26131535 | 0.44 |
|
|
|
| chr20_-_1170970 | 0.44 |
ENSDART00000043218
|
ube2j1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr8_+_1052572 | 0.44 |
ENSDART00000081445
|
ubox5
|
U-box domain containing 5 |
| chr5_-_3103278 | 0.44 |
|
|
|
| chr1_-_36039634 | 0.43 |
ENSDART00000148386
|
prmt9
|
protein arginine methyltransferase 9 |
| chr22_+_2014232 | 0.42 |
ENSDART00000165168
|
znf1157
|
zinc finger protein 1157 |
| chr4_+_17653548 | 0.41 |
ENSDART00000026509
|
cwf19l1
|
CWF19-like 1, cell cycle control |
| chr22_+_1484719 | 0.41 |
ENSDART00000159973
|
zgc:173486
|
zgc:173486 |
| chr7_+_69212144 | 0.41 |
ENSDART00000159799
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr12_+_23870021 | 0.40 |
ENSDART00000153136
|
psme4b
|
proteasome activator subunit 4b |
| chr1_-_45972732 | 0.40 |
ENSDART00000134450
|
ebpl
|
emopamil binding protein-like |
| chr15_-_35354406 | 0.40 |
ENSDART00000124780
|
agfg1a
|
ArfGAP with FG repeats 1a |
| chr21_+_19612140 | 0.40 |
ENSDART00000048581
|
fgf10a
|
fibroblast growth factor 10a |
| chr22_+_32274436 | 0.39 |
ENSDART00000092082
|
manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr23_-_31340025 | 0.39 |
ENSDART00000139746
|
si:dkey-261l7.2
|
si:dkey-261l7.2 |
| chr17_+_30431395 | 0.37 |
ENSDART00000153939
|
lpin1
|
lipin 1 |
| chr22_-_4605581 | 0.35 |
ENSDART00000028634
|
cherp
|
calcium homeostasis endoplasmic reticulum protein |
| chr16_+_8067699 | 0.35 |
ENSDART00000126041
|
ano10a
|
anoctamin 10a |
| chr13_-_36440277 | 0.35 |
ENSDART00000030133
|
synj2bp
|
synaptojanin 2 binding protein |
| chr3_+_22853476 | 0.35 |
ENSDART00000043190
|
lsm12a
|
LSM12 homolog a |
| chr18_+_7595039 | 0.35 |
ENSDART00000062143
|
zgc:77650
|
zgc:77650 |
| chr22_+_1624585 | 0.35 |
ENSDART00000159804
|
si:dkey-1b17.2
|
si:dkey-1b17.2 |
| chr2_-_6380292 | 0.34 |
ENSDART00000092182
|
ppm1la
|
protein phosphatase, Mg2+/Mn2+ dependent, 1La |
| chr5_+_59394192 | 0.34 |
|
|
|
| chr3_-_50424711 | 0.34 |
ENSDART00000047071
|
tmed1a
|
transmembrane p24 trafficking protein 1a |
| chr10_+_5688293 | 0.33 |
ENSDART00000172632
|
pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr25_+_16796766 | 0.33 |
|
|
|
| chr15_-_31296457 | 0.33 |
ENSDART00000008854
|
wsb1
|
WD repeat and SOCS box containing 1 |
| chr25_+_21001338 | 0.33 |
ENSDART00000017488
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr8_-_214434 | 0.33 |
ENSDART00000168412
|
hsd17b4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr13_-_36164801 | 0.32 |
ENSDART00000169768
|
slc8a3
|
solute carrier family 8 (sodium/calcium exchanger), member 3 |
| chr5_+_55607591 | 0.32 |
ENSDART00000148749
|
aatf
|
apoptosis antagonizing transcription factor |
| chr18_-_26131468 | 0.32 |
|
|
|
| chr22_-_10026610 | 0.32 |
ENSDART00000179409
|
si:ch211-222k6.1
|
si:ch211-222k6.1 |
| chr11_-_7146819 | 0.31 |
ENSDART00000172879
|
smim7
|
small integral membrane protein 7 |
| chr8_+_5222145 | 0.31 |
ENSDART00000035676
|
bnip3la
|
BCL2/adenovirus E1B interacting protein 3-like a |
| chr10_-_5846612 | 0.31 |
ENSDART00000161096
|
ankrd55
|
ankyrin repeat domain 55 |
| chr9_+_8920002 | 0.31 |
ENSDART00000132443
ENSDART00000139687 |
carkd
|
carbohydrate kinase domain containing |
| chr21_-_3462128 | 0.31 |
ENSDART00000139194
|
dym
|
dymeclin |
| chr19_-_29707065 | 0.31 |
ENSDART00000130815
ENSDART00000172590 ENSDART00000103437 |
e2f3
|
E2F transcription factor 3 |
| chr3_-_50424836 | 0.31 |
ENSDART00000047071
|
tmed1a
|
transmembrane p24 trafficking protein 1a |
| chr6_+_13273286 | 0.30 |
ENSDART00000032331
|
gmppab
|
GDP-mannose pyrophosphorylase Ab |
| chr13_+_36797201 | 0.30 |
ENSDART00000026313
|
tmx1
|
thioredoxin-related transmembrane protein 1 |
| chr10_+_21477579 | 0.30 |
ENSDART00000142447
|
etf1b
|
eukaryotic translation termination factor 1b |
| chr25_-_24150270 | 0.30 |
ENSDART00000073527
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr2_+_23004159 | 0.29 |
ENSDART00000123442
|
zgc:161973
|
zgc:161973 |
| chr2_-_11720585 | 0.28 |
ENSDART00000019392
|
sdr16c5a
|
short chain dehydrogenase/reductase family 16C, member 5a |
| chr14_-_46608393 | 0.28 |
ENSDART00000074038
|
scyl1
|
SCY1-like, kinase-like 1 |
| chr22_+_2231377 | 0.28 |
ENSDART00000143366
|
si:dkey-4c15.8
|
si:dkey-4c15.8 |
| chr14_-_3937426 | 0.27 |
ENSDART00000169527
|
snx25
|
sorting nexin 25 |
| chr21_-_31215400 | 0.27 |
ENSDART00000121946
|
crcp
|
calcitonin gene-related peptide-receptor component protein |
| chr17_+_9934584 | 0.27 |
ENSDART00000169522
ENSDART00000160156 |
sec23a
|
Sec23 homolog A, COPII coat complex component |
| chr23_+_33792005 | 0.27 |
ENSDART00000024695
|
dazap2
|
DAZ associated protein 2 |
| chr22_-_680961 | 0.27 |
ENSDART00000149320
|
arl8a
|
ADP-ribosylation factor-like 8A |
| chr23_+_7758140 | 0.27 |
ENSDART00000164255
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr5_-_56254422 | 0.26 |
ENSDART00000014028
|
ppm1db
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Db |
| chr3_+_32539409 | 0.26 |
ENSDART00000136292
|
rnf25
|
ring finger protein 25 |
| chr21_-_28579951 | 0.26 |
|
|
|
| chr11_+_36117256 | 0.26 |
ENSDART00000141529
|
atxn7l2a
|
ataxin 7-like 2a |
| chr15_+_47207251 | 0.25 |
ENSDART00000154481
|
stard10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr9_+_54736764 | 0.24 |
ENSDART00000148928
|
rbm27
|
RNA binding motif protein 27 |
| chr13_-_36164510 | 0.24 |
ENSDART00000169768
|
slc8a3
|
solute carrier family 8 (sodium/calcium exchanger), member 3 |
| chr6_-_1638631 | 0.23 |
ENSDART00000087039
|
ENSDARG00000015563
|
ENSDARG00000015563 |
| chr20_+_54544931 | 0.23 |
|
|
|
| chr15_+_17164373 | 0.23 |
ENSDART00000123197
ENSDART00000154418 |
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr4_+_344355 | 0.23 |
ENSDART00000142225
ENSDART00000163436 |
tmem181
|
transmembrane protein 181 |
| chr15_+_47207100 | 0.23 |
ENSDART00000154481
|
stard10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr4_+_16798009 | 0.22 |
ENSDART00000039027
|
golt1ba
|
golgi transport 1Ba |
| chr8_-_14201234 | 0.22 |
ENSDART00000090371
|
sin3b
|
SIN3 transcription regulator family member B |
| chr7_-_44623926 | 0.21 |
ENSDART00000073735
|
rrad
|
Ras-related associated with diabetes |
| chr10_+_21477307 | 0.21 |
ENSDART00000064558
|
etf1b
|
eukaryotic translation termination factor 1b |
| chr19_+_1148544 | 0.21 |
ENSDART00000166088
|
zgc:63863
|
zgc:63863 |
| chr7_-_73620443 | 0.20 |
ENSDART00000158891
ENSDART00000111622 |
FP236812.3
|
ENSDARG00000079652 |
| chr1_-_22466834 | 0.20 |
ENSDART00000144208
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr2_+_1139993 | 0.20 |
|
|
|
| chr25_+_15983777 | 0.20 |
ENSDART00000165598
ENSDART00000061753 |
far1
|
fatty acyl CoA reductase 1 |
| chr23_-_7283589 | 0.20 |
ENSDART00000156369
|
CR589876.1
|
ENSDARG00000096997 |
| chr6_-_1638559 | 0.20 |
ENSDART00000087039
|
ENSDARG00000015563
|
ENSDARG00000015563 |
| chr22_+_2644250 | 0.19 |
ENSDART00000031485
|
znf1162
|
zinc finger protein 1162 |
| chr20_+_17507402 | 0.19 |
|
|
|
| chr3_+_36474929 | 0.18 |
ENSDART00000158284
|
pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr25_-_12538613 | 0.18 |
ENSDART00000162463
|
zcchc14
|
zinc finger, CCHC domain containing 14 |
| chr20_+_54762145 | 0.18 |
ENSDART00000166592
|
ENSDARG00000102965
|
ENSDARG00000102965 |
| chr21_+_40393503 | 0.17 |
ENSDART00000171244
|
ssh2b
|
slingshot protein phosphatase 2b |
| chr18_+_20493291 | 0.17 |
ENSDART00000100668
|
kbtbd4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr20_+_22900165 | 0.17 |
ENSDART00000058527
|
scfd2
|
sec1 family domain containing 2 |
| chr9_+_33450418 | 0.16 |
ENSDART00000122803
|
usp9
|
ubiquitin specific peptidase 9 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.1 | GO:0072319 | clathrin coat disassembly(GO:0072318) vesicle uncoating(GO:0072319) |
| 0.6 | 1.7 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.5 | 2.0 | GO:0046098 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine metabolic process(GO:0046098) guanine biosynthetic process(GO:0046099) hypoxanthine metabolic process(GO:0046100) |
| 0.4 | 2.5 | GO:0000730 | DNA recombinase assembly(GO:0000730) |
| 0.4 | 1.6 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.4 | 0.7 | GO:0046048 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.3 | 2.3 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.3 | 2.3 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.3 | 1.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.3 | 1.1 | GO:0003400 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.2 | 2.2 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.2 | 0.7 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.2 | 2.9 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.2 | 0.6 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.1 | 1.1 | GO:0009109 | coenzyme catabolic process(GO:0009109) lipid particle organization(GO:0034389) |
| 0.1 | 0.6 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.3 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 2.0 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 0.6 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 1.8 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 0.9 | GO:0090148 | membrane fission(GO:0090148) |
| 0.1 | 0.6 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 1.0 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.1 | 2.8 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.1 | 1.0 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 0.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.5 | GO:0097345 | positive regulation of mitochondrial membrane permeability(GO:0035794) mitochondrial outer membrane permeabilization(GO:0097345) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 0.0 | 0.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 1.6 | GO:0035138 | pectoral fin morphogenesis(GO:0035138) |
| 0.0 | 0.2 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.3 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 1.6 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.0 | 1.7 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 0.8 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.4 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.9 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 2.1 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 1.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.2 | GO:0043931 | ossification involved in bone maturation(GO:0043931) bone maturation(GO:0070977) |
| 0.0 | 1.0 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.4 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.4 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.6 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.2 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.3 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.5 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.3 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.3 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 2.1 | GO:0016197 | endosomal transport(GO:0016197) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.6 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 2.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.3 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.8 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.4 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 4.6 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.6 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 2.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.8 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 1.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0017125 | deoxycytidyl transferase activity(GO:0017125) |
| 0.7 | 2.0 | GO:0052657 | guanine phosphoribosyltransferase activity(GO:0052657) |
| 0.5 | 1.6 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.4 | 1.7 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.3 | 1.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.3 | 1.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 0.7 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.2 | 0.7 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.2 | 0.6 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.2 | 1.6 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.4 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 0.3 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 0.6 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.3 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) amidine-lyase activity(GO:0016842) |
| 0.1 | 0.5 | GO:0097001 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.1 | 0.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.3 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.2 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.1 | 0.4 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 0.4 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.3 | GO:0046922 | peptide-O-fucosyltransferase activity(GO:0046922) |
| 0.1 | 1.8 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 4.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 4.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 2.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.6 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0042171 | lysophospholipase activity(GO:0004622) lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 2.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.8 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.8 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 2.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.8 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 1.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 4.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 0.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 3.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 3.1 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 0.5 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |