Project
DANIO-CODE
Navigation
Downloads
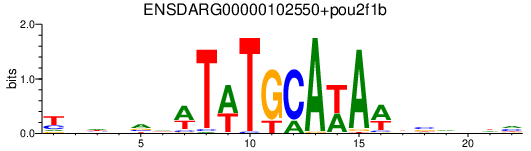
Results for ENSDARG00000102550+pou2f1b
Z-value: 0.57
Transcription factors associated with ENSDARG00000102550+pou2f1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou2f1b
|
ENSDARG00000007996 | POU class 2 homeobox 1b |
|
ENSDARG00000102550
|
ENSDARG00000102550 | ENSDARG00000102550 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CABZ01080370.1 | dr10_dc_chr15_-_61472_61544 | 0.64 | 7.1e-03 | Click! |
| pou2f1b | dr10_dc_chr9_+_34510091_34510171 | 0.43 | 9.4e-02 | Click! |
Activity profile of ENSDARG00000102550+pou2f1b motif
Sorted Z-values of ENSDARG00000102550+pou2f1b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ENSDARG00000102550+pou2f1b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_42095643 | 3.76 |
ENSDART00000100967
|
si:dkeyp-114g9.1
|
si:dkeyp-114g9.1 |
| chr12_+_22438815 | 1.44 |
ENSDART00000172066
|
polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr16_+_52961277 | 1.24 |
ENSDART00000163151
|
cep72
|
centrosomal protein 72 |
| chr22_+_24112851 | 1.18 |
|
|
|
| chr7_-_26226451 | 1.09 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr19_+_4835526 | 0.90 |
|
|
|
| chr11_+_30400284 | 0.84 |
ENSDART00000169833
|
gb:eh507706
|
expressed sequence EH507706 |
| chr22_+_24291633 | 0.83 |
ENSDART00000165618
|
ccdc50
|
coiled-coil domain containing 50 |
| chr12_+_22438908 | 0.82 |
ENSDART00000172066
|
polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr14_-_46980330 | 0.81 |
ENSDART00000043751
ENSDART00000141357 |
macrod1
|
MACRO domain containing 1 |
| chr13_+_33331767 | 0.77 |
ENSDART00000177841
|
zgc:136302
|
zgc:136302 |
| chr21_+_10609580 | 0.76 |
ENSDART00000102304
|
lman1
|
lectin, mannose-binding, 1 |
| chr16_+_52961219 | 0.75 |
ENSDART00000163151
|
cep72
|
centrosomal protein 72 |
| chr17_+_16038358 | 0.74 |
ENSDART00000155336
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr17_+_1063519 | 0.74 |
|
|
|
| chr22_+_21593142 | 0.72 |
ENSDART00000133939
|
tle2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr1_-_54586004 | 0.72 |
ENSDART00000152687
|
si:ch211-286b5.4
|
si:ch211-286b5.4 |
| chr17_-_20264762 | 0.71 |
|
|
|
| chr4_+_1727494 | 0.70 |
ENSDART00000149448
|
AL929171.1
|
ENSDARG00000095920 |
| chr15_-_14102102 | 0.67 |
ENSDART00000139068
|
zgc:114130
|
zgc:114130 |
| chr22_-_10861268 | 0.63 |
ENSDART00000145229
|
arhgef18b
|
rho/rac guanine nucleotide exchange factor (GEF) 18b |
| chr10_-_25737220 | 0.62 |
ENSDART00000135058
|
sod1
|
superoxide dismutase 1, soluble |
| chr5_+_68960543 | 0.61 |
ENSDART00000169013
|
arl6ip4
|
ADP-ribosylation factor-like 6 interacting protein 4 |
| chr25_-_2932779 | 0.61 |
ENSDART00000149117
ENSDART00000137950 |
si:ch1073-296i8.2
|
si:ch1073-296i8.2 |
| chr5_-_47323502 | 0.58 |
ENSDART00000175026
|
ccnh
|
cyclin H |
| chr23_+_12225922 | 0.57 |
ENSDART00000136046
|
ppp1r3da
|
protein phosphatase 1, regulatory subunit 3Da |
| chr22_-_37899243 | 0.55 |
ENSDART00000076128
|
ppp1r2
|
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
| chr8_+_13753663 | 0.55 |
|
|
|
| chr23_-_36178206 | 0.52 |
ENSDART00000153852
ENSDART00000153206 ENSDART00000155040 ENSDART00000156899 ENSDART00000157077 ENSDART00000155063 |
BX005254.1
|
ENSDARG00000093797 |
| chr21_-_39501604 | 0.51 |
ENSDART00000006971
|
sept4a
|
septin 4a |
| chr2_-_21780380 | 0.51 |
ENSDART00000144587
|
plcd1b
|
phospholipase C, delta 1b |
| chr6_-_39524075 | 0.51 |
ENSDART00000065038
|
atf1
|
activating transcription factor 1 |
| chr7_-_32598812 | 0.51 |
|
|
|
| chr16_+_19831573 | 0.50 |
ENSDART00000135359
|
macc1
|
metastasis associated in colon cancer 1 |
| chr12_-_22116694 | 0.50 |
ENSDART00000038310
ENSDART00000132731 |
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr21_+_30684976 | 0.49 |
ENSDART00000040443
|
zgc:110224
|
zgc:110224 |
| chr12_+_26943407 | 0.49 |
ENSDART00000153054
|
fbrs
|
fibrosin |
| chr8_-_16479425 | 0.49 |
ENSDART00000146469
ENSDART00000132681 |
ttc39a
|
tetratricopeptide repeat domain 39A |
| chr18_+_8959686 | 0.49 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr16_+_53632152 | 0.48 |
ENSDART00000124691
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr12_-_26943743 | 0.47 |
|
|
|
| chr7_-_26226924 | 0.46 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr18_+_2250457 | 0.46 |
|
|
|
| chr17_+_23291331 | 0.46 |
ENSDART00000128073
|
ppp1r3ca
|
protein phosphatase 1, regulatory subunit 3Ca |
| chr4_-_15604719 | 0.45 |
ENSDART00000122520
ENSDART00000041420 ENSDART00000162356 |
chchd3a
|
coiled-coil-helix-coiled-coil-helix domain containing 3a |
| chr3_-_59690168 | 0.45 |
ENSDART00000035878
|
cdr2l
|
cerebellar degeneration-related protein 2-like |
| KN150460v1_-_33517 | 0.44 |
|
|
|
| chr5_-_64983964 | 0.44 |
ENSDART00000147707
|
camsap1b
|
calmodulin regulated spectrin-associated protein 1b |
| chr16_-_43107682 | 0.44 |
ENSDART00000142003
|
nudt17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chr16_+_34092695 | 0.44 |
ENSDART00000058424
|
fam46ba
|
family with sequence similarity 46, member Ba |
| KN150307v1_+_6450 | 0.43 |
ENSDART00000159959
|
CABZ01038161.1
|
ENSDARG00000098784 |
| chr17_-_25813136 | 0.43 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr4_-_3340315 | 0.43 |
ENSDART00000009076
|
pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr13_+_18376540 | 0.42 |
ENSDART00000142622
|
si:ch211-198a12.6
|
si:ch211-198a12.6 |
| chr7_+_20092235 | 0.41 |
ENSDART00000139274
|
ponzr1
|
plac8 onzin related protein 1 |
| chr10_-_40897835 | 0.41 |
ENSDART00000076316
|
zgc:113625
|
zgc:113625 |
| chr7_+_22521055 | 0.40 |
ENSDART00000146801
|
rbm4.3
|
RNA binding motif protein 4.3 |
| chr16_+_53703721 | 0.40 |
ENSDART00000083534
|
gpatch3
|
G patch domain containing 3 |
| chr14_-_3937426 | 0.40 |
ENSDART00000169527
|
snx25
|
sorting nexin 25 |
| chr25_-_25339397 | 0.40 |
ENSDART00000171589
|
hrasa
|
v-Ha-ras Harvey rat sarcoma viral oncogene homolog a |
| chr9_-_11884101 | 0.39 |
ENSDART00000146731
ENSDART00000134553 |
AL935144.1
|
ENSDARG00000093040 |
| chr23_-_33848571 | 0.39 |
ENSDART00000027959
|
racgap1
|
Rac GTPase activating protein 1 |
| chr6_+_49724436 | 0.39 |
ENSDART00000154738
|
stx16
|
syntaxin 16 |
| chr14_+_30390987 | 0.38 |
|
|
|
| chr15_-_18273640 | 0.38 |
ENSDART00000141508
|
btr16
|
bloodthirsty-related gene family, member 16 |
| chr1_-_54488824 | 0.38 |
ENSDART00000150430
|
pane1
|
proliferation associated nuclear element |
| chr22_+_14811628 | 0.38 |
ENSDART00000122740
|
gtpbp1l
|
GTP binding protein 1, like |
| chr16_-_6298105 | 0.37 |
|
|
|
| chr5_-_47323448 | 0.37 |
ENSDART00000175026
|
ccnh
|
cyclin H |
| chr19_-_26188710 | 0.37 |
ENSDART00000052393
|
pard6gb
|
par-6 family cell polarity regulator gamma b |
| chr19_-_29707065 | 0.37 |
ENSDART00000130815
ENSDART00000172590 ENSDART00000103437 |
e2f3
|
E2F transcription factor 3 |
| chr3_-_50424711 | 0.36 |
ENSDART00000047071
|
tmed1a
|
transmembrane p24 trafficking protein 1a |
| chr20_-_9135267 | 0.36 |
ENSDART00000125133
|
mysm1
|
Myb-like, SWIRM and MPN domains 1 |
| chr4_-_4603205 | 0.35 |
ENSDART00000130601
|
CABZ01020840.1
|
ENSDARG00000090401 |
| chr16_-_44979403 | 0.35 |
|
|
|
| chr9_+_52308527 | 0.35 |
|
|
|
| chr11_+_5819952 | 0.35 |
ENSDART00000126084
|
ctdspl3
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 3 |
| chr7_-_24373258 | 0.35 |
ENSDART00000172282
|
rgp1
|
GP1 homolog, RAB6A GEF complex partner 1 |
| chr19_+_2942092 | 0.34 |
ENSDART00000177848
|
CABZ01066434.1
|
ENSDARG00000107451 |
| chr1_-_50395003 | 0.34 |
ENSDART00000035150
|
spast
|
spastin |
| chr16_+_34092822 | 0.34 |
ENSDART00000058424
|
fam46ba
|
family with sequence similarity 46, member Ba |
| chr13_+_12651744 | 0.34 |
ENSDART00000090000
|
si:ch211-233a24.2
|
si:ch211-233a24.2 |
| chr22_-_12601276 | 0.33 |
ENSDART00000143261
|
cep70
|
centrosomal protein 70 |
| chr24_-_20496410 | 0.32 |
ENSDART00000166135
|
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr19_-_40111964 | 0.32 |
|
|
|
| chr17_-_7194149 | 0.32 |
ENSDART00000098731
|
stxbp5b
|
syntaxin binding protein 5b (tomosyn) |
| chr13_-_23626319 | 0.32 |
ENSDART00000124152
|
rgs17
|
regulator of G protein signaling 17 |
| chr16_+_53703883 | 0.31 |
ENSDART00000083534
|
gpatch3
|
G patch domain containing 3 |
| chr15_+_29460803 | 0.31 |
ENSDART00000155198
|
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr7_-_39467720 | 0.31 |
ENSDART00000052201
|
ccdc96
|
coiled-coil domain containing 96 |
| chr7_+_19348229 | 0.31 |
ENSDART00000007310
ENSDART00000166355 |
zgc:171731
|
zgc:171731 |
| chr20_+_19093849 | 0.30 |
ENSDART00000025509
|
tdh
|
L-threonine dehydrogenase |
| chr20_-_42095478 | 0.30 |
ENSDART00000100967
|
si:dkeyp-114g9.1
|
si:dkeyp-114g9.1 |
| chr20_-_34851706 | 0.30 |
ENSDART00000148066
|
znf395b
|
zinc finger protein 395b |
| chr14_+_35065189 | 0.30 |
ENSDART00000171565
|
zbtb3
|
zinc finger and BTB domain containing 3 |
| chr9_+_2523927 | 0.30 |
ENSDART00000166326
|
si:ch73-167c12.2
|
si:ch73-167c12.2 |
| chr22_+_2575708 | 0.29 |
|
|
|
| chr4_+_830826 | 0.29 |
|
|
|
| chr25_-_2932820 | 0.29 |
ENSDART00000149117
ENSDART00000137950 |
si:ch1073-296i8.2
|
si:ch1073-296i8.2 |
| chr2_+_9762627 | 0.28 |
ENSDART00000003465
|
gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr10_-_244746 | 0.28 |
ENSDART00000136551
|
klhl35
|
kelch-like family member 35 |
| chr13_+_27638410 | 0.27 |
ENSDART00000159281
|
asrgl1
|
asparaginase like 1 |
| chr22_-_10861329 | 0.27 |
ENSDART00000145229
|
arhgef18b
|
rho/rac guanine nucleotide exchange factor (GEF) 18b |
| chr10_+_34057791 | 0.26 |
ENSDART00000149934
|
kl
|
klotho |
| chr14_-_16448507 | 0.26 |
ENSDART00000001159
|
mgat4b
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B |
| chr24_-_20496308 | 0.26 |
ENSDART00000166135
|
zbtb47b
|
zinc finger and BTB domain containing 47b |
| KN149781v1_+_5174 | 0.26 |
ENSDART00000165339
|
CDC37
|
cell division cycle 37 |
| chr7_-_24373522 | 0.26 |
ENSDART00000048921
|
rgp1
|
GP1 homolog, RAB6A GEF complex partner 1 |
| chr4_+_5823740 | 0.26 |
ENSDART00000121743
|
si:ch73-352p4.5
|
si:ch73-352p4.5 |
| chr11_-_21243422 | 0.26 |
ENSDART00000080116
|
ikbke
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr7_-_26226628 | 0.26 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr24_-_36307897 | 0.25 |
ENSDART00000154395
|
esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr7_-_24373315 | 0.25 |
ENSDART00000048921
|
rgp1
|
GP1 homolog, RAB6A GEF complex partner 1 |
| chr7_+_22520928 | 0.25 |
ENSDART00000146801
|
rbm4.3
|
RNA binding motif protein 4.3 |
| chr3_+_19095975 | 0.25 |
ENSDART00000134514
|
smarca4a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4a |
| chr4_-_10770022 | 0.24 |
|
|
|
| chr15_+_34734212 | 0.24 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr21_+_34085267 | 0.24 |
ENSDART00000024750
|
hmgb3b
|
high mobility group box 3b |
| chr25_+_2968769 | 0.24 |
|
|
|
| chr23_+_16880876 | 0.24 |
ENSDART00000141966
|
zgc:114081
|
zgc:114081 |
| chr19_-_31212648 | 0.24 |
ENSDART00000125893
ENSDART00000145581 |
trit1
|
tRNA isopentenyltransferase 1 |
| chr23_+_12226078 | 0.23 |
ENSDART00000136046
|
ppp1r3da
|
protein phosphatase 1, regulatory subunit 3Da |
| chr19_-_29706969 | 0.23 |
ENSDART00000130815
ENSDART00000172590 ENSDART00000103437 |
e2f3
|
E2F transcription factor 3 |
| chr3_+_36970806 | 0.23 |
ENSDART00000055228
|
psmc3ip
|
PSMC3 interacting protein |
| chr21_-_42813859 | 0.22 |
|
|
|
| chr12_-_10343382 | 0.22 |
ENSDART00000152788
|
mki67
|
marker of proliferation Ki-67 |
| chr15_-_34799968 | 0.22 |
ENSDART00000110964
|
bag6
|
BCL2-associated athanogene 6 |
| chr2_-_40067309 | 0.22 |
|
|
|
| chr21_+_30685020 | 0.21 |
ENSDART00000040443
|
zgc:110224
|
zgc:110224 |
| chr20_-_15045029 | 0.21 |
ENSDART00000152775
|
mettl13
|
methyltransferase like 13 |
| chr12_-_35481361 | 0.21 |
ENSDART00000158658
ENSDART00000168958 ENSDART00000162175 |
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr3_-_50424836 | 0.21 |
ENSDART00000047071
|
tmed1a
|
transmembrane p24 trafficking protein 1a |
| chr4_-_4603294 | 0.21 |
ENSDART00000130601
|
CABZ01020840.1
|
ENSDARG00000090401 |
| chr4_+_76572863 | 0.21 |
ENSDART00000156692
|
arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr23_-_77607 | 0.21 |
|
|
|
| chr1_-_18980902 | 0.20 |
ENSDART00000129853
|
apela
|
apelin receptor early endogenous ligand |
| chr16_-_26201059 | 0.20 |
ENSDART00000148653
ENSDART00000148923 |
tmem145
|
transmembrane protein 145 |
| chr3_+_19515578 | 0.20 |
ENSDART00000007857
|
mettl2a
|
methyltransferase like 2A |
| chr2_-_44545564 | 0.20 |
ENSDART00000146192
|
tmem55ba
|
transmembrane protein 55Ba |
| chr16_-_42866318 | 0.20 |
ENSDART00000176570
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| KN150307v1_+_6577 | 0.19 |
ENSDART00000159959
|
CABZ01038161.1
|
ENSDARG00000098784 |
| chr5_-_66589204 | 0.19 |
ENSDART00000125874
|
zcchc8
|
zinc finger, CCHC domain containing 8 |
| chr20_+_46588659 | 0.19 |
ENSDART00000060695
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr7_-_6226607 | 0.19 |
ENSDART00000129239
|
HIST2H2AB (1 of many)
|
histone cluster 2 H2A family member b |
| chr21_-_34891991 | 0.19 |
ENSDART00000065337
|
kif20a
|
kinesin family member 20A |
| chr2_-_16690351 | 0.19 |
ENSDART00000057216
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr7_+_28782292 | 0.19 |
ENSDART00000137241
|
fam60al
|
family with sequence similarity 60, member A, like |
| chr12_-_36113358 | 0.19 |
|
|
|
| chr23_-_2957169 | 0.18 |
ENSDART00000165955
|
zhx3
|
zinc fingers and homeoboxes 3 |
| chr5_+_44205128 | 0.18 |
ENSDART00000136965
|
ctsla
|
cathepsin La |
| chr7_+_24373948 | 0.18 |
ENSDART00000150233
|
gba2
|
glucosidase, beta (bile acid) 2 |
| chr5_-_1432791 | 0.18 |
ENSDART00000128274
|
golga2
|
golgin A2 |
| chr4_-_5822882 | 0.18 |
ENSDART00000008898
|
foxm1
|
forkhead box M1 |
| chr10_+_26636293 | 0.18 |
ENSDART00000079187
|
fhl1b
|
four and a half LIM domains 1b |
| chr22_+_24576319 | 0.18 |
ENSDART00000164256
|
ENSDARG00000100186
|
ENSDARG00000100186 |
| chr21_-_14719281 | 0.16 |
ENSDART00000067004
|
phpt1
|
phosphohistidine phosphatase 1 |
| chr13_+_13799208 | 0.16 |
ENSDART00000079154
|
rpia
|
ribose 5-phosphate isomerase A (ribose 5-phosphate epimerase) |
| chr22_+_36562719 | 0.16 |
ENSDART00000178537
|
CABZ01045213.1
|
ENSDARG00000107720 |
| chr11_+_5522377 | 0.16 |
ENSDART00000013203
|
cse1l
|
CSE1 chromosome segregation 1-like (yeast) |
| chr4_-_75732882 | 0.16 |
|
|
|
| chr20_-_35567985 | 0.15 |
ENSDART00000016090
|
pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr19_+_1743359 | 0.15 |
ENSDART00000166744
|
dennd3a
|
DENN/MADD domain containing 3a |
| chr5_-_64983812 | 0.15 |
ENSDART00000147707
|
camsap1b
|
calmodulin regulated spectrin-associated protein 1b |
| chr8_-_40412363 | 0.15 |
ENSDART00000161142
|
tbc1d10aa
|
TBC1 domain family, member 10Aa |
| chr20_+_46588401 | 0.15 |
ENSDART00000060695
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr19_-_40598818 | 0.15 |
ENSDART00000150972
|
efcab1
|
EF-hand calcium binding domain 1 |
| chr4_-_2370333 | 0.14 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr21_+_11274670 | 0.14 |
ENSDART00000091835
|
gtf3c5
|
general transcription factor IIIC, polypeptide 5 |
| chr6_+_49724271 | 0.14 |
ENSDART00000002693
|
stx16
|
syntaxin 16 |
| chr2_-_44545661 | 0.14 |
ENSDART00000146192
|
tmem55ba
|
transmembrane protein 55Ba |
| chr12_+_28710843 | 0.14 |
ENSDART00000133703
|
cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr15_-_43447148 | 0.13 |
ENSDART00000077386
|
prss16
|
protease, serine, 16 (thymus) |
| chr4_+_20074260 | 0.13 |
ENSDART00000024925
|
gcc1
|
GRIP and coiled-coil domain containing 1 |
| chr16_-_42866236 | 0.13 |
ENSDART00000112879
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr1_+_22583970 | 0.13 |
ENSDART00000036448
|
lias
|
lipoic acid synthetase |
| chr7_+_28782951 | 0.13 |
|
|
|
| chr2_-_22875246 | 0.13 |
ENSDART00000159641
|
znf644a
|
zinc finger protein 644a |
| chr13_+_18376642 | 0.12 |
ENSDART00000142622
|
si:ch211-198a12.6
|
si:ch211-198a12.6 |
| chr4_-_3015097 | 0.12 |
ENSDART00000128934
ENSDART00000019518 |
aebp2
|
AE binding protein 2 |
| chr9_+_12473243 | 0.12 |
ENSDART00000132709
|
tmem41aa
|
transmembrane protein 41aa |
| chr3_-_26830876 | 0.12 |
|
|
|
| chr22_+_14811881 | 0.12 |
ENSDART00000122740
|
gtpbp1l
|
GTP binding protein 1, like |
| chr22_-_29938022 | 0.11 |
|
|
|
| chr6_+_29420518 | 0.11 |
ENSDART00000065293
|
usp13
|
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr6_+_28127363 | 0.11 |
|
|
|
| chr19_-_41882385 | 0.11 |
|
|
|
| chr4_+_5332964 | 0.10 |
ENSDART00000123375
|
zgc:113263
|
zgc:113263 |
| chr19_+_9313687 | 0.10 |
ENSDART00000136957
|
kmt2ba
|
lysine (K)-specific methyltransferase 2Ba |
| chr14_-_47211080 | 0.09 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr16_-_52961072 | 0.09 |
|
|
|
| chr6_+_41098443 | 0.09 |
ENSDART00000143741
|
fkbp5
|
FK506 binding protein 5 |
| chr13_+_28339823 | 0.09 |
|
|
|
| chr24_-_36307855 | 0.09 |
ENSDART00000111891
|
esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr4_+_11763234 | 0.09 |
ENSDART00000153714
ENSDART00000156950 |
BX465862.3
|
ENSDARG00000096099 |
| chr1_-_38097153 | 0.09 |
ENSDART00000149080
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr5_-_12706568 | 0.08 |
ENSDART00000051666
|
ppm1f
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr7_-_41232765 | 0.08 |
ENSDART00000173577
|
ENSDARG00000105669
|
ENSDARG00000105669 |
| chr21_-_14719149 | 0.08 |
ENSDART00000067004
|
phpt1
|
phosphohistidine phosphatase 1 |
| chr7_+_5783441 | 0.08 |
ENSDART00000167099
|
CU459186.5
|
Histone H3.2 |
| chr6_-_3837266 | 0.08 |
ENSDART00000059212
|
unc50
|
unc-50 homolog (C. elegans) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0039531 | negative regulation of type I interferon production(GO:0032480) cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) cellular response to virus(GO:0098586) |
| 0.2 | 2.0 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.2 | 0.5 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.4 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 0.7 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.6 | GO:0071451 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 0.3 | GO:0043243 | positive regulation of protein complex disassembly(GO:0043243) |
| 0.1 | 0.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 1.3 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 0.2 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.3 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.2 | GO:0044706 | adult heart development(GO:0007512) female pregnancy(GO:0007565) embryo implantation(GO:0007566) multi-multicellular organism process(GO:0044706) apelin receptor signaling pathway(GO:0060183) trophoblast cell migration(GO:0061450) regulation of trophoblast cell migration(GO:1901163) positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 0.3 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.1 | 0.2 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.1 | 0.6 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 0.3 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.1 | 0.2 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.1 | 0.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.4 | GO:0072078 | renal tubule morphogenesis(GO:0061333) nephron tubule morphogenesis(GO:0072078) nephron epithelium morphogenesis(GO:0072088) |
| 0.0 | 0.9 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.5 | GO:0072178 | pronephric duct morphogenesis(GO:0039023) nephric duct morphogenesis(GO:0072178) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 1.9 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.8 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.4 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.2 | GO:0007589 | body fluid secretion(GO:0007589) |
| 0.0 | 0.3 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.5 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.4 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.4 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.1 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.1 | GO:0001715 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) ectodermal cell fate specification(GO:0001715) ectodermal cell differentiation(GO:0010668) regulation of ectodermal cell fate specification(GO:0042665) regulation of ectoderm development(GO:2000383) |
| 0.0 | 0.1 | GO:0090200 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.8 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.2 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.0 | 0.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.3 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.2 | GO:1990798 | pancreas regeneration(GO:1990798) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.2 | 0.8 | GO:0034066 | RIC1-RGP1 guanyl-nucleotide exchange factor complex(GO:0034066) |
| 0.2 | 0.5 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 2.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.4 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 1.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.6 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 0.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.5 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 1.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.5 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.1 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.4 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.6 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.3 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.1 | 0.3 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 1.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.4 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.2 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 0.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.4 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.0 | 0.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.4 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.8 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.9 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.0 | 0.6 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 2.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 1.2 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.3 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |