Project
DANIO-CODE
Navigation
Downloads
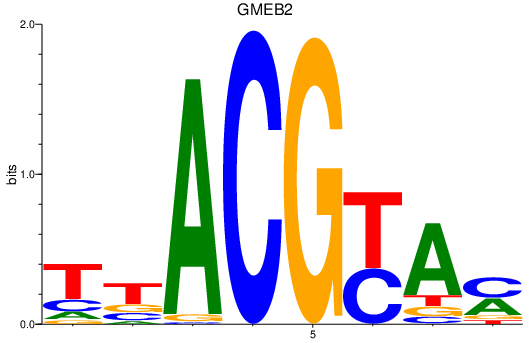
Results for GMEB2
Z-value: 2.10
Transcription factors associated with GMEB2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GMEB2
|
ENSDARG00000093240 | si_ch73-302a13.2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GMEB2 | dr10_dc_chr23_-_41929027_41929077 | -0.77 | 4.9e-04 | Click! |
Activity profile of GMEB2 motif
Sorted Z-values of GMEB2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GMEB2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_25555313 | 7.67 |
ENSDART00000021672
|
epc2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr11_-_2437396 | 6.20 |
|
|
|
| chr3_-_29779725 | 5.36 |
ENSDART00000151501
|
RUNDC1
|
RUN domain containing 1 |
| chr3_-_29779598 | 5.05 |
ENSDART00000151501
|
RUNDC1
|
RUN domain containing 1 |
| chr18_-_20684934 | 4.98 |
ENSDART00000005145
|
asz1
|
ankyrin repeat, SAM and basic leucine zipper domain containing 1 |
| chr24_+_34183462 | 4.69 |
ENSDART00000143995
|
zgc:92591
|
zgc:92591 |
| chr23_-_35691369 | 4.56 |
ENSDART00000142369
|
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr2_-_17443607 | 4.32 |
ENSDART00000136207
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr9_-_25555345 | 4.21 |
ENSDART00000021672
|
epc2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr25_+_21001126 | 4.05 |
ENSDART00000079012
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr12_-_28679813 | 3.80 |
ENSDART00000020667
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr2_-_17443642 | 3.66 |
ENSDART00000136207
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr10_+_29251234 | 3.64 |
ENSDART00000123033
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr24_+_34183557 | 3.62 |
ENSDART00000143995
|
zgc:92591
|
zgc:92591 |
| chr9_+_25109716 | 3.50 |
ENSDART00000037025
|
slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr15_+_5125138 | 3.45 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr4_+_18525469 | 3.28 |
ENSDART00000154154
|
BX649398.1
|
ENSDARG00000097195 |
| chr19_-_44459352 | 3.26 |
ENSDART00000151084
|
uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr11_-_2437361 | 3.21 |
|
|
|
| chr7_-_37624410 | 3.18 |
ENSDART00000084282
|
papd5
|
PAP associated domain containing 5 |
| chr4_+_9010972 | 3.18 |
ENSDART00000058007
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr13_+_36554762 | 3.14 |
ENSDART00000136030
|
atp5s
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit s |
| chr3_-_29779552 | 3.14 |
ENSDART00000151501
|
RUNDC1
|
RUN domain containing 1 |
| chr15_+_5124897 | 3.12 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr5_-_23092592 | 3.12 |
ENSDART00000024815
|
fam76b
|
family with sequence similarity 76, member B |
| chr7_-_37624329 | 3.09 |
ENSDART00000084282
|
papd5
|
PAP associated domain containing 5 |
| chr15_+_5124690 | 3.07 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr9_-_25555518 | 2.99 |
ENSDART00000021672
|
epc2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr5_-_29818204 | 2.92 |
ENSDART00000098285
|
atf5a
|
activating transcription factor 5a |
| chr18_-_14910497 | 2.89 |
ENSDART00000099701
|
selo
|
selenoprotein O |
| chr18_-_20684687 | 2.86 |
ENSDART00000005145
|
asz1
|
ankyrin repeat, SAM and basic leucine zipper domain containing 1 |
| chr11_-_36212977 | 2.84 |
ENSDART00000165203
|
usp48
|
ubiquitin specific peptidase 48 |
| chr24_-_42053682 | 2.77 |
ENSDART00000169725
|
zbtb14
|
zinc finger and BTB domain containing 14 |
| chr6_-_52677363 | 2.71 |
ENSDART00000083830
|
sdc4
|
syndecan 4 |
| chr10_+_16543480 | 2.64 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr20_-_31840932 | 2.64 |
ENSDART00000137679
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr3_+_22204475 | 2.60 |
ENSDART00000055676
|
ENSDARG00000038186
|
ENSDARG00000038186 |
| chr24_-_23853204 | 2.60 |
ENSDART00000130053
|
zmp:0000000991
|
zmp:0000000991 |
| chr23_+_17596504 | 2.59 |
ENSDART00000002714
|
slc17a9b
|
solute carrier family 17 (vesicular nucleotide transporter), member 9b |
| chr16_-_33864015 | 2.54 |
|
|
|
| chr14_+_10319183 | 2.50 |
ENSDART00000136480
|
nup62l
|
nucleoporin 62 like |
| chr16_-_5205600 | 2.47 |
ENSDART00000148955
|
bckdhb
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr13_+_42183685 | 2.47 |
ENSDART00000158367
|
ide
|
insulin-degrading enzyme |
| chr13_+_29904945 | 2.46 |
ENSDART00000057525
|
dnajb12a
|
DnaJ (Hsp40) homolog, subfamily B, member 12a |
| chr17_-_51729568 | 2.44 |
ENSDART00000157244
|
exd2
|
exonuclease 3'-5' domain containing 2 |
| chr17_-_51729707 | 2.38 |
ENSDART00000157244
|
exd2
|
exonuclease 3'-5' domain containing 2 |
| chr5_+_6005049 | 2.35 |
|
|
|
| chr8_-_37989317 | 2.35 |
ENSDART00000067809
|
rab11fip1a
|
RAB11 family interacting protein 1 (class I) a |
| chr5_+_25538153 | 2.33 |
ENSDART00000010041
|
dhfr
|
dihydrofolate reductase |
| chr23_+_38313746 | 2.31 |
ENSDART00000129593
|
znf217
|
zinc finger protein 217 |
| chr11_+_11136919 | 2.30 |
ENSDART00000026135
|
ly75
|
lymphocyte antigen 75 |
| chr1_+_48770997 | 2.28 |
ENSDART00000015007
|
taf5
|
TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr8_-_37989415 | 2.28 |
ENSDART00000067809
|
rab11fip1a
|
RAB11 family interacting protein 1 (class I) a |
| chr1_-_25369230 | 2.25 |
ENSDART00000171752
|
pdcd4a
|
programmed cell death 4a |
| chr20_+_25587099 | 2.20 |
ENSDART00000063052
|
hook1
|
hook microtubule-tethering protein 1 |
| chr7_+_13130111 | 2.18 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr17_-_43409443 | 2.17 |
ENSDART00000156418
|
itpk1b
|
inositol-tetrakisphosphate 1-kinase b |
| chr6_-_23953873 | 2.15 |
ENSDART00000165697
|
ENSDARG00000098123
|
ENSDARG00000098123 |
| chr10_+_29251158 | 2.13 |
ENSDART00000123033
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr7_+_13130177 | 2.10 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr21_+_6291936 | 2.02 |
ENSDART00000136539
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr8_+_30103357 | 1.99 |
ENSDART00000133717
|
fancc
|
Fanconi anemia, complementation group C |
| chr13_+_3807780 | 1.98 |
ENSDART00000012759
|
yipf3
|
Yip1 domain family, member 3 |
| chr17_-_25813136 | 1.96 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr7_+_41033439 | 1.96 |
ENSDART00000016660
|
ENSDARG00000016964
|
ENSDARG00000016964 |
| chr22_+_35229352 | 1.95 |
ENSDART00000061315
ENSDART00000146430 |
tsc22d2
|
TSC22 domain family 2 |
| chr13_+_42183490 | 1.91 |
ENSDART00000158367
|
ide
|
insulin-degrading enzyme |
| chr20_-_38030337 | 1.91 |
ENSDART00000153451
|
angel2
|
angel homolog 2 (Drosophila) |
| chr7_+_69290790 | 1.88 |
ENSDART00000109507
|
RAP1GDS1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr20_+_54545050 | 1.87 |
|
|
|
| chr7_-_23623141 | 1.85 |
|
|
|
| KN150712v1_-_26150 | 1.82 |
|
|
|
| chr25_+_21001016 | 1.81 |
ENSDART00000079012
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr15_-_20476476 | 1.79 |
ENSDART00000008589
|
chp2
|
calcineurin-like EF-hand protein 2 |
| chr6_-_10492724 | 1.73 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr3_+_22204569 | 1.73 |
ENSDART00000055676
|
ENSDARG00000038186
|
ENSDARG00000038186 |
| chr20_-_9212139 | 1.71 |
ENSDART00000064140
|
ylpm1
|
YLP motif containing 1 |
| chr17_-_36913302 | 1.70 |
ENSDART00000154981
|
senp6b
|
SUMO1/sentrin specific peptidase 6b |
| chr15_+_5124616 | 1.70 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr16_+_14311427 | 1.68 |
ENSDART00000113679
|
dap3
|
death associated protein 3 |
| chr12_+_17481598 | 1.68 |
ENSDART00000170449
ENSDART00000111565 |
pms2
|
PMS1 homolog 2, mismatch repair system component |
| chr15_+_5124820 | 1.67 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr25_+_21001338 | 1.66 |
ENSDART00000017488
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr5_-_53748764 | 1.60 |
ENSDART00000160781
ENSDART00000160965 ENSDART00000161692 |
zmynd19
|
zinc finger, MYND-type containing 19 |
| chr14_+_8641470 | 1.59 |
ENSDART00000158025
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr18_-_14910429 | 1.59 |
ENSDART00000099701
|
selo
|
selenoprotein O |
| chr13_+_29904913 | 1.57 |
ENSDART00000057525
|
dnajb12a
|
DnaJ (Hsp40) homolog, subfamily B, member 12a |
| chr10_+_16543577 | 1.54 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr17_+_16082983 | 1.49 |
ENSDART00000133154
|
znf395a
|
zinc finger protein 395a |
| chr17_+_22050088 | 1.48 |
|
|
|
| chr25_+_35540123 | 1.48 |
ENSDART00000126326
|
rpgrip1l
|
RPGRIP1-like |
| chr5_-_56539892 | 1.48 |
|
|
|
| chr22_+_35229139 | 1.46 |
ENSDART00000061315
ENSDART00000146430 |
tsc22d2
|
TSC22 domain family 2 |
| chr5_+_56539999 | 1.46 |
ENSDART00000167660
|
pja2
|
praja ring finger 2 |
| chr20_-_32502517 | 1.43 |
ENSDART00000153411
|
afg1lb
|
AFG1 like ATPase b |
| chr7_+_56423315 | 1.42 |
|
|
|
| chr14_-_44794907 | 1.41 |
ENSDART00000172805
|
CABZ01074913.1
|
ENSDARG00000105401 |
| chr1_+_22583970 | 1.39 |
ENSDART00000036448
|
lias
|
lipoic acid synthetase |
| chr9_-_23431846 | 1.39 |
ENSDART00000053282
|
ccnt2a
|
cyclin T2a |
| chr14_+_8641516 | 1.38 |
ENSDART00000158025
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr13_-_36554590 | 1.38 |
ENSDART00000085298
|
l2hgdh
|
L-2-hydroxyglutarate dehydrogenase |
| chr2_-_32502787 | 1.36 |
ENSDART00000110821
|
ttc19
|
tetratricopeptide repeat domain 19 |
| chr19_+_46667326 | 1.35 |
ENSDART00000158032
|
grinaa
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1a (glutamate binding) |
| chr22_-_22277418 | 1.35 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr22_-_22277386 | 1.35 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr9_-_7676688 | 1.35 |
ENSDART00000102706
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr25_-_20968390 | 1.33 |
ENSDART00000109520
|
fbxl14a
|
F-box and leucine-rich repeat protein 14a |
| chr20_+_25586940 | 1.28 |
ENSDART00000063052
|
hook1
|
hook microtubule-tethering protein 1 |
| chr5_+_44054942 | 1.24 |
ENSDART00000136642
|
dapk1
|
death-associated protein kinase 1 |
| chr2_+_32842978 | 1.22 |
ENSDART00000143127
|
si:dkey-154p10.3
|
si:dkey-154p10.3 |
| chr1_+_36593187 | 1.21 |
ENSDART00000171340
|
fam193a
|
family with sequence similarity 193, member A |
| chr6_-_40457472 | 1.19 |
ENSDART00000103878
|
vhl
|
von Hippel-Lindau tumor suppressor |
| chr3_-_48458042 | 1.18 |
ENSDART00000156822
|
cdip1
|
cell death-inducing p53 target 1 |
| chr3_+_32802124 | 1.17 |
ENSDART00000112742
|
nbr1b
|
neighbor of brca1 gene 1b |
| chr12_-_28841710 | 1.16 |
ENSDART00000142933
|
znf646
|
zinc finger protein 646 |
| chr3_+_32657833 | 1.15 |
ENSDART00000171895
|
tbc1d10b
|
TBC1 domain family, member 10b |
| chr21_+_1622040 | 1.10 |
ENSDART00000148540
|
fech
|
ferrochelatase |
| chr1_-_25369054 | 1.10 |
ENSDART00000171752
|
pdcd4a
|
programmed cell death 4a |
| chr19_-_12058542 | 1.06 |
ENSDART00000168308
|
si:ch73-49k18.1
|
si:ch73-49k18.1 |
| chr16_+_41004516 | 1.06 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr10_+_16544000 | 1.06 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr11_+_43127567 | 1.04 |
ENSDART00000172982
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr18_+_5303006 | 1.03 |
|
|
|
| chr18_+_49253587 | 1.03 |
ENSDART00000059285
|
yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr23_+_38314255 | 1.00 |
ENSDART00000129593
|
znf217
|
zinc finger protein 217 |
| chr12_+_20290791 | 0.97 |
ENSDART00000115015
|
arhgap17a
|
Rho GTPase activating protein 17a |
| chr14_-_44794961 | 0.97 |
ENSDART00000172805
|
CABZ01074913.1
|
ENSDARG00000105401 |
| chr16_+_40181425 | 0.93 |
ENSDART00000155421
|
cenpw
|
centromere protein W |
| chr7_-_20378231 | 0.91 |
|
|
|
| chr6_-_52677210 | 0.91 |
ENSDART00000083830
|
sdc4
|
syndecan 4 |
| chr8_-_1154220 | 0.91 |
ENSDART00000149969
ENSDART00000016800 |
znf367
|
zinc finger protein 367 |
| chr6_-_51541332 | 0.89 |
ENSDART00000156336
ENSDART00000154512 |
si:dkey-6e2.2
|
si:dkey-6e2.2 |
| chr16_+_14311271 | 0.87 |
ENSDART00000113679
|
dap3
|
death associated protein 3 |
| chr17_-_6294655 | 0.87 |
ENSDART00000064700
|
fuca2
|
fucosidase, alpha-L- 2, plasma |
| chr6_-_39905773 | 0.86 |
ENSDART00000148612
|
tsfm
|
Ts translation elongation factor, mitochondrial |
| chr12_-_28841396 | 0.85 |
|
|
|
| chr21_+_4345107 | 0.85 |
ENSDART00000025612
|
phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr13_+_3808095 | 0.83 |
ENSDART00000012759
|
yipf3
|
Yip1 domain family, member 3 |
| chr9_+_22846535 | 0.79 |
ENSDART00000101765
|
si:dkey-189g17.2
|
si:dkey-189g17.2 |
| chr9_+_19719018 | 0.78 |
ENSDART00000129934
ENSDART00000125416 |
pknox1.1
|
pbx/knotted 1 homeobox 1.1 |
| chr18_-_14910358 | 0.78 |
ENSDART00000099701
|
selo
|
selenoprotein O |
| chr19_+_40439263 | 0.76 |
ENSDART00000017359
|
sfpq
|
splicing factor proline/glutamine-rich |
| chr20_-_9212089 | 0.75 |
ENSDART00000064140
|
ylpm1
|
YLP motif containing 1 |
| chr1_+_5464124 | 0.74 |
ENSDART00000144559
|
cnppd1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr3_+_32801936 | 0.74 |
ENSDART00000112742
|
nbr1b
|
neighbor of brca1 gene 1b |
| chr17_-_5918853 | 0.74 |
ENSDART00000058890
ENSDART00000171084 |
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr8_-_30935167 | 0.72 |
ENSDART00000049944
|
smarcb1a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1a |
| chr8_+_20423036 | 0.72 |
ENSDART00000079618
|
rexo1
|
REX1, RNA exonuclease 1 homolog |
| chr1_+_48770878 | 0.71 |
ENSDART00000015007
|
taf5
|
TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr25_-_20968316 | 0.69 |
ENSDART00000109520
|
fbxl14a
|
F-box and leucine-rich repeat protein 14a |
| chr7_-_20378183 | 0.67 |
|
|
|
| chr7_-_23623275 | 0.65 |
ENSDART00000123203
|
vbp1
|
von Hippel-Lindau binding protein 1 |
| chr6_+_8927621 | 0.63 |
ENSDART00000157552
|
zgc:112023
|
zgc:112023 |
| chr13_-_12353513 | 0.63 |
ENSDART00000134132
|
lamtor3
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 |
| chr20_+_38934332 | 0.62 |
ENSDART00000061334
|
ift172
|
intraflagellar transport 172 |
| chr7_+_13130367 | 0.61 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr14_+_10319051 | 0.60 |
ENSDART00000136480
|
nup62l
|
nucleoporin 62 like |
| chr24_+_34183407 | 0.60 |
ENSDART00000143995
|
zgc:92591
|
zgc:92591 |
| chr14_+_8641354 | 0.59 |
ENSDART00000047993
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr3_+_12966604 | 0.58 |
ENSDART00000172460
|
PRKAR1B (1 of many)
|
protein kinase cAMP-dependent type I regulatory subunit beta |
| chr16_-_30775394 | 0.56 |
|
|
|
| chr16_+_20488737 | 0.55 |
ENSDART00000059619
|
fkbp14
|
FK506 binding protein 14 |
| chr9_+_26029780 | 0.49 |
ENSDART00000127135
|
gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr9_+_23238634 | 0.48 |
ENSDART00000012478
|
mmadhc
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria |
| chr6_-_37767593 | 0.42 |
ENSDART00000078316
|
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr6_-_6925121 | 0.42 |
ENSDART00000081760
|
ihhb
|
Indian hedgehog homolog b |
| chr15_+_32940372 | 0.41 |
ENSDART00000170098
|
spg20b
|
spastic paraplegia 20b (Troyer syndrome) |
| chr2_+_21431575 | 0.40 |
ENSDART00000109568
|
pip4k2ab
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha b |
| chr12_+_28841405 | 0.40 |
ENSDART00000033878
|
znf668
|
zinc finger protein 668 |
| chr7_+_52486748 | 0.36 |
ENSDART00000111444
|
ppip5k1a
|
diphosphoinositol pentakisphosphate kinase 1a |
| chr25_-_28225408 | 0.36 |
ENSDART00000126490
|
fnbp4
|
formin binding protein 4 |
| chr2_+_32843040 | 0.36 |
ENSDART00000143127
|
si:dkey-154p10.3
|
si:dkey-154p10.3 |
| chr7_+_41033728 | 0.36 |
ENSDART00000016660
|
ENSDARG00000016964
|
ENSDARG00000016964 |
| chr16_-_20488542 | 0.35 |
ENSDART00000059623
|
plekha8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr17_-_37247455 | 0.35 |
ENSDART00000108514
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr1_+_25110899 | 0.34 |
ENSDART00000129471
|
gucy1b3
|
guanylate cyclase 1, soluble, beta 3 |
| chr14_+_37205204 | 0.34 |
ENSDART00000173192
|
pcdh1b
|
protocadherin 1b |
| chr13_+_3807844 | 0.32 |
ENSDART00000012759
|
yipf3
|
Yip1 domain family, member 3 |
| chr9_+_22008242 | 0.30 |
ENSDART00000136902
|
txndc9
|
thioredoxin domain containing 9 |
| chr2_-_42279072 | 0.27 |
ENSDART00000047055
|
trim55a
|
tripartite motif containing 55a |
| chr25_-_6262259 | 0.27 |
ENSDART00000083539
|
sin3aa
|
SIN3 transcription regulator family member Aa |
| chr14_-_21635094 | 0.27 |
ENSDART00000054420
|
rad9a
|
RAD9 checkpoint clamp component A |
| chr7_+_13130332 | 0.26 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr24_+_5176362 | 0.23 |
ENSDART00000155926
|
si:ch73-206p6.1
|
si:ch73-206p6.1 |
| chr16_-_21683623 | 0.23 |
ENSDART00000146914
|
mpp6a
|
membrane protein, palmitoylated 6a (MAGUK p55 subfamily member 6) |
| chr16_+_41065107 | 0.20 |
ENSDART00000058586
|
dek
|
DEK proto-oncogene |
| chr5_-_23092453 | 0.16 |
ENSDART00000024815
|
fam76b
|
family with sequence similarity 76, member B |
| chr12_+_11611780 | 0.15 |
ENSDART00000150191
|
waplb
|
WAPL cohesin release factor b |
| chr7_+_41033650 | 0.15 |
ENSDART00000016660
|
ENSDARG00000016964
|
ENSDARG00000016964 |
| KN150109v1_-_3702 | 0.15 |
ENSDART00000175695
|
CABZ01089385.1
|
ENSDARG00000107509 |
| chr25_-_28225449 | 0.14 |
ENSDART00000126490
|
fnbp4
|
formin binding protein 4 |
| chr7_-_67093071 | 0.11 |
ENSDART00000159408
|
psmd7
|
proteasome 26S subunit, non-ATPase 7 |
| chr20_-_36905932 | 0.08 |
ENSDART00000166966
|
slc25a27
|
solute carrier family 25, member 27 |
| chr8_-_1154004 | 0.08 |
ENSDART00000149969
ENSDART00000016800 |
znf367
|
zinc finger protein 367 |
| chr24_-_37596370 | 0.07 |
ENSDART00000162538
|
cluap1
|
clusterin associated protein 1 |
| chr12_-_3630622 | 0.07 |
ENSDART00000164707
|
coa3a
|
cytochrome C oxidase assembly factor 3a |
| chr6_-_26905146 | 0.06 |
ENSDART00000011863
|
hdlbpa
|
high density lipoprotein binding protein a |
| chr7_-_39480446 | 0.06 |
ENSDART00000016803
|
grpel1
|
GrpE-like 1, mitochondrial |
| chr6_+_42478384 | 0.05 |
ENSDART00000084667
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr25_+_21001071 | 0.04 |
ENSDART00000079012
|
rad52
|
RAD52 homolog, DNA repair protein |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 7.6 | GO:0045002 | DNA recombinase assembly(GO:0000730) double-strand break repair via single-strand annealing(GO:0045002) |
| 1.1 | 3.2 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 1.0 | 5.1 | GO:0099542 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.9 | 3.6 | GO:0036445 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 0.8 | 2.3 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.7 | 8.0 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.6 | 14.9 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.6 | 5.3 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.6 | 7.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.4 | 4.4 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.4 | 1.2 | GO:0043576 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.4 | 3.5 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.4 | 1.5 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.3 | 3.6 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.3 | 2.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.3 | 1.0 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.2 | 0.9 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.2 | 1.4 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.2 | 1.9 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.2 | 4.0 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.2 | 5.2 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.2 | 1.9 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.2 | 13.0 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.1 | 1.5 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 1.3 | GO:0007584 | response to nutrient(GO:0007584) |
| 0.1 | 0.4 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 | 1.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 1.4 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.1 | 0.8 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.1 | 3.0 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 3.1 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 1.7 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 2.0 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 1.0 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 0.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.2 | GO:0050857 | regulation of antigen receptor-mediated signaling pathway(GO:0050854) regulation of B cell receptor signaling pathway(GO:0050855) positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 1.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.7 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 0.1 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 4.3 | GO:0015698 | inorganic anion transport(GO:0015698) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 2.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 3.9 | GO:0045055 | regulated exocytosis(GO:0045055) |
| 0.0 | 0.8 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.3 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 2.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.2 | GO:0071922 | regulation of sister chromatid cohesion(GO:0007063) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0021547 | midbrain-hindbrain boundary initiation(GO:0021547) |
| 0.0 | 1.4 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 2.6 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.0 | 1.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.7 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.6 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) response to amino acid(GO:0043200) cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 2.8 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 1.7 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 1.4 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 1.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.7 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 1.2 | GO:0010942 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 14.9 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 1.3 | 5.3 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 1.1 | 7.8 | GO:0071546 | pi-body(GO:0071546) |
| 0.9 | 4.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.7 | 3.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.6 | 1.7 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.5 | 3.2 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.5 | 1.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.4 | 3.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.4 | 1.9 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.4 | 3.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.3 | 1.3 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.2 | 2.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 1.9 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 8.0 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 3.0 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 1.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 1.2 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.1 | 2.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 8.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.6 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 1.9 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 0.7 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 5.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.6 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 3.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 2.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 2.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.3 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.9 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 1.2 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 8.0 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.9 | 5.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.9 | 5.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.7 | 5.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.7 | 13.0 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.4 | 1.7 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.4 | 1.5 | GO:0052725 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.3 | 14.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.3 | 3.6 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.3 | 4.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.2 | 3.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 3.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.2 | 0.7 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.2 | 1.7 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.2 | 1.4 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 3.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 1.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 3.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 7.0 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.1 | 0.4 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 2.3 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 9.1 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 4.3 | GO:0015103 | inorganic anion transmembrane transporter activity(GO:0015103) |
| 0.0 | 0.4 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 2.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 2.0 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 4.5 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 0.7 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 1.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.7 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 1.0 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.4 | GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors(GO:0016614) |
| 0.0 | 0.9 | GO:0042803 | protein homodimerization activity(GO:0042803) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 5.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 2.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 7.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 2.6 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 1.2 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 1.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 2.0 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 1.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.4 | 7.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.3 | 2.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.3 | 5.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.2 | 3.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 1.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.4 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.3 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.6 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |