Project
DANIO-CODE
Navigation
Downloads
Results for LHX3+lhx4
Z-value: 0.95
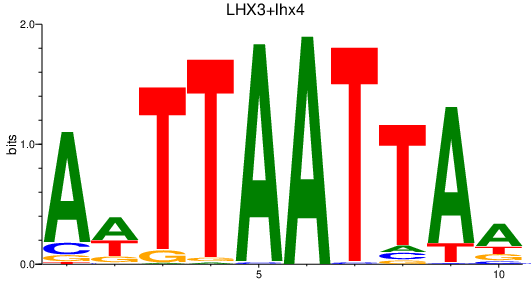
Transcription factors associated with LHX3+lhx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LHX3
|
ENSDARG00000003803 | LIM homeobox 3 |
|
lhx4
|
ENSDARG00000039458 | LIM homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lhx4 | dr10_dc_chr8_-_14446883_14446894 | 0.94 | 9.4e-08 | Click! |
| LHX3 | dr10_dc_chr5_+_71028018_71028045 | 0.93 | 1.3e-07 | Click! |
Activity profile of LHX3+lhx4 motif
Sorted Z-values of LHX3+lhx4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of LHX3+lhx4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_34832049 | 3.31 |
ENSDART00000100735
ENSDART00000133996 |
shox
|
short stature homeobox |
| chr13_+_24148599 | 3.19 |
ENSDART00000058629
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr20_-_22576513 | 2.81 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr8_-_34077387 | 2.70 |
ENSDART00000159208
ENSDART00000040126 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr16_+_23516127 | 2.62 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr4_+_9668755 | 2.48 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr20_+_19613133 | 2.34 |
ENSDART00000152548
ENSDART00000063696 |
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr17_+_15425559 | 2.15 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr20_-_3970778 | 2.15 |
ENSDART00000178724
ENSDART00000178565 |
TRIM67
|
tripartite motif containing 67 |
| chr21_+_25199691 | 2.13 |
ENSDART00000168140
ENSDART00000112783 |
tmem45b
|
transmembrane protein 45B |
| chr9_+_36092350 | 2.11 |
ENSDART00000005086
|
atp1a1b
|
ATPase, Na+/K+ transporting, alpha 1b polypeptide |
| chr16_+_46145286 | 2.05 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr19_-_5781358 | 2.03 |
ENSDART00000082050
|
zgc:174904
|
zgc:174904 |
| chr5_-_40894631 | 1.96 |
ENSDART00000121840
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr6_-_16590883 | 1.95 |
ENSDART00000153552
|
nomo
|
nodal modulator |
| chr14_-_26238386 | 1.88 |
ENSDART00000105933
|
tgfbi
|
transforming growth factor, beta-induced |
| chr17_+_38528709 | 1.87 |
ENSDART00000123298
|
stard9
|
StAR-related lipid transfer (START) domain containing 9 |
| chr24_-_1155215 | 1.80 |
ENSDART00000177356
|
itgb1a
|
integrin, beta 1a |
| chr14_-_20760787 | 1.79 |
|
|
|
| chr20_-_48677794 | 1.79 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr4_-_73485204 | 1.74 |
ENSDART00000122898
|
phf21b
|
PHD finger protein 21B |
| chr1_-_4757890 | 1.72 |
ENSDART00000114035
|
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr17_-_29102320 | 1.65 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr1_-_44476084 | 1.63 |
ENSDART00000034549
|
zgc:111983
|
zgc:111983 |
| chr14_+_47326080 | 1.61 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr19_-_39048324 | 1.59 |
ENSDART00000086717
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr23_+_36023748 | 1.57 |
|
|
|
| chr3_-_23444371 | 1.53 |
ENSDART00000087726
|
igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr20_-_9107294 | 1.53 |
ENSDART00000140792
|
OMA1
|
OMA1 zinc metallopeptidase |
| chr18_-_43890514 | 1.50 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr19_-_23037220 | 1.50 |
ENSDART00000090669
|
pleca
|
plectin a |
| chr6_-_18891908 | 1.50 |
ENSDART00000074327
|
igfbp2a
|
insulin-like growth factor binding protein 2a |
| chr6_+_23787804 | 1.48 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr10_-_43924675 | 1.47 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr7_+_26357875 | 1.47 |
ENSDART00000101044
|
hsbp1a
|
heat shock factor binding protein 1a |
| chr9_+_308260 | 1.46 |
ENSDART00000163474
|
stac3
|
SH3 and cysteine rich domain 3 |
| chr15_-_23441268 | 1.45 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr4_-_15442828 | 1.43 |
ENSDART00000157414
|
plxna4
|
plexin A4 |
| chr13_+_29640492 | 1.42 |
ENSDART00000160944
|
pax2a
|
paired box 2a |
| chr7_-_25623974 | 1.40 |
ENSDART00000173602
|
cd99l2
|
CD99 molecule-like 2 |
| chr11_+_30416115 | 1.38 |
ENSDART00000161662
|
ttbk1a
|
tau tubulin kinase 1a |
| chr24_-_6048914 | 1.37 |
ENSDART00000146830
ENSDART00000021981 |
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr14_-_17257773 | 1.36 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor-like 1a |
| chr1_+_21037251 | 1.36 |
|
|
|
| chr9_-_37940101 | 1.35 |
ENSDART00000087663
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr19_+_12995955 | 1.34 |
ENSDART00000132892
|
cthrc1a
|
collagen triple helix repeat containing 1a |
| chr18_+_21419735 | 1.34 |
ENSDART00000144523
|
necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr15_+_9351511 | 1.33 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr2_+_29729161 | 1.33 |
ENSDART00000141666
|
AL845362.1
|
ENSDARG00000093205 |
| chr22_+_16509286 | 1.33 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr24_-_14447773 | 1.33 |
|
|
|
| chr1_-_40208544 | 1.33 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr1_-_40208469 | 1.32 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr8_+_2428689 | 1.31 |
ENSDART00000081325
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr6_-_14821305 | 1.31 |
|
|
|
| chr18_+_43041488 | 1.30 |
|
|
|
| chr5_+_62987426 | 1.30 |
ENSDART00000178937
|
dnm1b
|
dynamin 1b |
| chr6_-_8075384 | 1.30 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr5_-_67233396 | 1.29 |
ENSDART00000051833
ENSDART00000124890 |
gsx1
|
GS homeobox 1 |
| chr11_+_7518953 | 1.29 |
ENSDART00000171813
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr7_-_39170016 | 1.27 |
ENSDART00000161191
|
BX842701.1
|
ENSDARG00000101872 |
| chr1_-_29919280 | 1.25 |
ENSDART00000152175
|
MZT1
|
mitotic spindle organizing protein 1 |
| chr24_+_35899507 | 1.25 |
ENSDART00000122408
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr19_+_5562107 | 1.22 |
ENSDART00000082080
|
jupb
|
junction plakoglobin b |
| chr22_+_38220826 | 1.22 |
ENSDART00000010537
|
cp
|
ceruloplasmin |
| chr7_+_25052687 | 1.21 |
ENSDART00000110347
|
cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr21_+_28441951 | 1.21 |
ENSDART00000077887
|
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr2_+_36718808 | 1.21 |
ENSDART00000002510
|
golim4b
|
golgi integral membrane protein 4b |
| chr17_+_25500291 | 1.20 |
ENSDART00000041721
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr14_+_2969679 | 1.19 |
ENSDART00000044678
|
ENSDARG00000034011
|
ENSDARG00000034011 |
| chr20_-_28739099 | 1.19 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr25_+_5695630 | 1.18 |
ENSDART00000161035
|
C12orf75
|
chromosome 12 open reading frame 75 |
| chr19_+_44185325 | 1.16 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr20_-_9474672 | 1.14 |
ENSDART00000152674
|
ENSDARG00000033201
|
ENSDARG00000033201 |
| chr7_-_24428781 | 1.12 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr8_+_43334181 | 1.11 |
ENSDART00000038566
|
fam101a
|
family with sequence similarity 101, member A |
| chr13_-_29290894 | 1.11 |
ENSDART00000150228
|
chata
|
choline O-acetyltransferase a |
| chr1_-_53403731 | 1.09 |
ENSDART00000171722
|
smdt1b
|
single-pass membrane protein with aspartate-rich tail 1b |
| chr1_+_39844690 | 1.09 |
ENSDART00000122059
|
scoca
|
short coiled-coil protein a |
| chr19_+_20183210 | 1.09 |
ENSDART00000164968
|
hoxa4a
|
homeobox A4a |
| chr17_+_25463178 | 1.08 |
ENSDART00000139451
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr1_-_674449 | 1.07 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr16_+_46435014 | 1.07 |
ENSDART00000144000
|
rpz2
|
rapunzel 2 |
| chr15_-_15513719 | 1.07 |
ENSDART00000101918
|
proca1
|
protein interacting with cyclin A1 |
| chr24_-_26165778 | 1.06 |
ENSDART00000080113
|
apodb
|
apolipoprotein Db |
| chr7_-_47900199 | 1.06 |
ENSDART00000174262
|
mtss1lb
|
metastasis suppressor 1-like b |
| chr14_-_16791447 | 1.05 |
ENSDART00000158002
|
CR812832.1
|
ENSDARG00000103278 |
| chr16_-_13733759 | 1.05 |
ENSDART00000164344
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr7_+_24762755 | 1.04 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr14_-_7774262 | 1.04 |
ENSDART00000045109
|
zgc:92242
|
zgc:92242 |
| chr13_-_29875809 | 1.03 |
ENSDART00000049624
|
wnt8b
|
wingless-type MMTV integration site family, member 8b |
| chr16_-_28943421 | 1.02 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr11_-_6051096 | 1.02 |
ENSDART00000147761
|
vsg1
|
vessel-specific 1 |
| chr23_+_5632124 | 1.01 |
ENSDART00000059307
|
smpd2a
|
sphingomyelin phosphodiesterase 2a, neutral membrane (neutral sphingomyelinase) |
| chr8_+_23464087 | 1.01 |
ENSDART00000026316
|
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr10_+_13042496 | 1.00 |
ENSDART00000158919
|
lpar1
|
lysophosphatidic acid receptor 1 |
| chr12_-_4497094 | 0.99 |
ENSDART00000163651
|
trpm4b.2
|
transient receptor potential cation channel, subfamily M, member 4b, transient receptor potential cation channel, subfamily M, member 4b, tandem duplicate 2 |
| chr15_-_14616083 | 0.99 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr21_+_10775209 | 0.99 |
|
|
|
| chr2_+_21267793 | 0.98 |
ENSDART00000099913
|
pla2g4aa
|
phospholipase A2, group IVAa (cytosolic, calcium-dependent) |
| chr1_-_18809429 | 0.97 |
ENSDART00000124260
|
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr9_-_14137664 | 0.96 |
ENSDART00000132747
|
prkag3b
|
protein kinase, AMP-activated, gamma 3b non-catalytic subunit |
| chr1_-_30801520 | 0.95 |
ENSDART00000007770
|
lbx1b
|
ladybird homeobox 1b |
| chr6_+_52350428 | 0.95 |
ENSDART00000151612
|
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr14_+_27936303 | 0.95 |
ENSDART00000003293
|
mid2
|
midline 2 |
| chr5_-_13738619 | 0.95 |
ENSDART00000090788
|
npffr1l2
|
neuropeptide FF receptor 1 like 2 |
| chr25_-_21718782 | 0.94 |
ENSDART00000175132
|
ENSDARG00000051873
|
ENSDARG00000051873 |
| chr13_-_9554501 | 0.94 |
ENSDART00000139541
|
ENSDARG00000069828
|
ENSDARG00000069828 |
| chr1_-_22160662 | 0.94 |
ENSDART00000054386
|
qdprb1
|
quinoid dihydropteridine reductase b1 |
| chr9_+_947223 | 0.93 |
ENSDART00000159892
|
dock9a
|
dedicator of cytokinesis 9a |
| chr9_-_51975919 | 0.93 |
ENSDART00000006612
|
tbr1b
|
T-box, brain, 1b |
| chr5_-_40894693 | 0.92 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr20_-_21031504 | 0.92 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr3_-_19218660 | 0.92 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr25_-_5883620 | 0.92 |
ENSDART00000136054
|
snx22
|
sorting nexin 22 |
| chr19_-_27284726 | 0.91 |
ENSDART00000089699
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr14_-_4038642 | 0.91 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr13_+_33151628 | 0.91 |
ENSDART00000135200
|
ccdc28b
|
coiled-coil domain containing 28B |
| chr10_-_27087472 | 0.90 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr8_-_11512545 | 0.90 |
ENSDART00000133932
|
si:ch211-248e11.2
|
si:ch211-248e11.2 |
| chr12_-_7902815 | 0.89 |
ENSDART00000088100
|
ank3b
|
ankyrin 3b |
| chr7_+_19243998 | 0.89 |
ENSDART00000173766
|
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr21_+_45802046 | 0.89 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr8_-_19166630 | 0.88 |
|
|
|
| chr19_-_28546417 | 0.87 |
ENSDART00000130922
ENSDART00000079114 |
irx1b
|
iroquois homeobox 1b |
| chr5_+_71028018 | 0.87 |
ENSDART00000164893
ENSDART00000159658 ENSDART00000097164 ENSDART00000124939 ENSDART00000171230 |
LHX3
|
LIM homeobox 3 |
| chr14_-_45883839 | 0.86 |
|
|
|
| chr8_-_17130805 | 0.86 |
|
|
|
| chr21_-_5692160 | 0.86 |
ENSDART00000075137
ENSDART00000151202 |
ccni
|
cyclin I |
| chr15_-_18226612 | 0.86 |
ENSDART00000012064
|
pih1d2
|
PIH1 domain containing 2 |
| chr16_-_28921444 | 0.86 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr16_-_45211650 | 0.86 |
ENSDART00000165186
|
si:dkey-33i11.9
|
si:dkey-33i11.9 |
| chr4_+_22759177 | 0.85 |
ENSDART00000146272
|
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr20_-_27193158 | 0.84 |
ENSDART00000062096
|
si:dkeyp-55f12.3
|
si:dkeyp-55f12.3 |
| chr23_+_36016450 | 0.84 |
ENSDART00000103035
|
hoxc6a
|
homeobox C6a |
| chr15_-_37949371 | 0.83 |
ENSDART00000031418
|
hsc70
|
heat shock cognate 70 |
| chr3_-_60930025 | 0.82 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr25_+_13309645 | 0.81 |
ENSDART00000010495
|
znrf1
|
zinc and ring finger 1 |
| chr14_-_47083485 | 0.80 |
ENSDART00000172512
|
elf2a
|
E74-like factor 2a (ets domain transcription factor) |
| chr15_-_8333117 | 0.80 |
ENSDART00000061351
|
tnfrsf19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr12_-_34621359 | 0.79 |
|
|
|
| chr21_-_25567978 | 0.79 |
ENSDART00000133134
|
efemp2b
|
EGF containing fibulin-like extracellular matrix protein 2b |
| chr4_-_935448 | 0.79 |
ENSDART00000171855
|
sim1b
|
single-minded family bHLH transcription factor 1b |
| chr20_-_54192057 | 0.77 |
ENSDART00000023550
|
hsp90aa1.2
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 2 |
| chr24_+_24686591 | 0.77 |
ENSDART00000080969
|
trim55b
|
tripartite motif containing 55b |
| chr8_-_12365342 | 0.76 |
ENSDART00000113286
|
phf19
|
PHD finger protein 19 |
| chr7_-_27877036 | 0.75 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr7_-_26331734 | 0.75 |
|
|
|
| chr11_+_15935769 | 0.74 |
ENSDART00000158824
|
ctbs
|
chitobiase, di-N-acetyl- |
| chr5_-_9162684 | 0.74 |
|
|
|
| chr3_-_43730993 | 0.72 |
|
|
|
| chr24_-_24306469 | 0.72 |
ENSDART00000154149
|
BX323067.1
|
ENSDARG00000097984 |
| chr13_+_24148474 | 0.71 |
ENSDART00000058629
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr10_-_11303185 | 0.70 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr24_+_37692365 | 0.70 |
|
|
|
| chr16_+_29060022 | 0.70 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr6_-_43679553 | 0.70 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr2_-_49114158 | 0.69 |
|
|
|
| chr7_+_21006803 | 0.68 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr20_-_2936289 | 0.68 |
ENSDART00000104667
|
cdk19
|
cyclin-dependent kinase 19 |
| chr24_-_24001766 | 0.68 |
ENSDART00000173052
ENSDART00000173220 |
map7d2b
|
MAP7 domain containing 2b |
| chr2_+_1825714 | 0.67 |
ENSDART00000148624
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr1_+_18842158 | 0.67 |
ENSDART00000103089
|
ENSDARG00000070320
|
ENSDARG00000070320 |
| chr4_+_5246465 | 0.67 |
ENSDART00000137966
|
ccdc167
|
coiled-coil domain containing 167 |
| chr2_+_29507819 | 0.66 |
|
|
|
| chr14_-_47129958 | 0.65 |
ENSDART00000172166
|
NDUFC1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr3_+_50511676 | 0.65 |
ENSDART00000102202
|
ppap2d
|
phosphatidic acid phosphatase type 2D |
| chr8_-_12365285 | 0.64 |
ENSDART00000142150
|
phf19
|
PHD finger protein 19 |
| chr8_+_31812204 | 0.64 |
ENSDART00000077053
|
plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr12_-_28248133 | 0.64 |
ENSDART00000016283
|
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr19_-_4079427 | 0.64 |
ENSDART00000170325
|
map7d1b
|
MAP7 domain containing 1b |
| chr3_+_32234028 | 0.64 |
ENSDART00000155967
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr14_-_1342450 | 0.64 |
ENSDART00000060417
|
cetn4
|
centrin 4 |
| chr1_+_40795366 | 0.63 |
ENSDART00000146310
|
dtx4
|
deltex 4, E3 ubiquitin ligase |
| chr16_-_13734068 | 0.63 |
ENSDART00000164344
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr5_-_37494209 | 0.63 |
ENSDART00000084890
|
si:ch211-284e13.4
|
si:ch211-284e13.4 |
| chr19_-_39048402 | 0.63 |
ENSDART00000086717
|
col16a1
|
collagen, type XVI, alpha 1 |
| KN150123v1_-_37214 | 0.62 |
|
|
|
| chr20_-_52526843 | 0.62 |
ENSDART00000132941
|
si:ch1073-287p18.1
|
si:ch1073-287p18.1 |
| chr24_+_24316486 | 0.62 |
ENSDART00000147658
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr4_+_70768190 | 0.61 |
|
|
|
| KN150474v1_-_163952 | 0.60 |
|
|
|
| chr6_+_28215039 | 0.60 |
ENSDART00000104394
|
smx5
|
smx5 |
| chr13_+_32609784 | 0.60 |
ENSDART00000160138
|
sobpa
|
sine oculis binding protein homolog (Drosophila) a |
| chr20_+_36098607 | 0.59 |
ENSDART00000021456
|
opn5
|
opsin 5 |
| chr7_+_19300351 | 0.59 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr16_-_26083913 | 0.59 |
ENSDART00000166681
|
frrs1l
|
ferric-chelate reductase 1-like |
| chr17_-_12812747 | 0.58 |
ENSDART00000022874
ENSDART00000046025 |
psma6a
|
proteasome subunit alpha 6a |
| chr15_-_21003820 | 0.58 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr16_-_21334269 | 0.58 |
ENSDART00000145837
|
si:dkey-271j15.3
|
si:dkey-271j15.3 |
| chr2_+_7054686 | 0.57 |
ENSDART00000057385
|
nos1apb
|
nitric oxide synthase 1 (neuronal) adaptor protein b |
| chr25_+_23326630 | 0.57 |
|
|
|
| chr21_+_27241416 | 0.57 |
|
|
|
| chr20_+_7596461 | 0.56 |
ENSDART00000127975
ENSDART00000132481 ENSDART00000144551 |
bloc1s2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr15_+_23615727 | 0.56 |
ENSDART00000152720
|
MARK4
|
microtubule affinity regulating kinase 4 |
| chr16_-_48189682 | 0.55 |
|
|
|
| chr14_-_31514534 | 0.54 |
ENSDART00000003345
|
arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr2_-_52792076 | 0.52 |
ENSDART00000097716
|
zgc:136336
|
zgc:136336 |
| chr3_-_32205873 | 0.52 |
ENSDART00000156918
|
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr25_+_23326562 | 0.52 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.6 | 2.3 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.5 | 1.5 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.5 | 2.3 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.4 | 1.2 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.4 | 1.5 | GO:0040014 | regulation of multicellular organism growth(GO:0040014) |
| 0.4 | 1.1 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.4 | 1.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.4 | 1.5 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) sarcoplasmic reticulum calcium ion transport(GO:0070296) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.3 | 1.0 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.3 | 1.0 | GO:0002706 | regulation of lymphocyte mediated immunity(GO:0002706) |
| 0.3 | 2.8 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.3 | 1.8 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.3 | 1.4 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.3 | 0.8 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.3 | 1.3 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.3 | 1.3 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.2 | 0.7 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.2 | 1.0 | GO:0042698 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) inflammatory response to wounding(GO:0090594) |
| 0.2 | 1.2 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.2 | 1.3 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.2 | 0.7 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.2 | 1.1 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.2 | 1.4 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.2 | 2.6 | GO:1900052 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.2 | 1.3 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.2 | 1.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 2.0 | GO:0043584 | nose development(GO:0043584) |
| 0.2 | 0.5 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.2 | 0.5 | GO:0009713 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.2 | 0.3 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.2 | 0.6 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.2 | 0.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 1.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 1.1 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.2 | 0.9 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.6 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.1 | 0.9 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 1.7 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 1.0 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 1.4 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 0.4 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 0.9 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.1 | 2.1 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.2 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.1 | 3.3 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.1 | 1.8 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.1 | 1.2 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.1 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.4 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 1.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.3 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 2.2 | GO:0015696 | ammonium transport(GO:0015696) |
| 0.1 | 0.6 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 1.1 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 0.8 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 1.2 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 0.6 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 0.2 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 5.0 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.1 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.3 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 3.0 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.4 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 1.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 1.3 | GO:0000302 | response to reactive oxygen species(GO:0000302) |
| 0.0 | 2.2 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 1.0 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.4 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.7 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.3 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 2.3 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 1.4 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.0 | 1.6 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.8 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 1.4 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.0 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.5 | GO:0048634 | regulation of muscle organ development(GO:0048634) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.6 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.1 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.0 | 0.2 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 1.8 | GO:0048738 | cardiac muscle tissue development(GO:0048738) |
| 0.0 | 2.2 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.4 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.9 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.6 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.6 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.3 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 1.5 | GO:0060026 | convergent extension(GO:0060026) |
| 0.0 | 0.2 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.5 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.4 | GO:0050821 | protein stabilization(GO:0050821) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 3.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.3 | 1.3 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.2 | 1.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.2 | 1.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 1.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 0.9 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 1.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.6 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.8 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 1.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.3 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 0.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 1.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 2.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 1.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 1.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.1 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.6 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.6 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 2.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 1.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 4.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.8 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 2.1 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 3.4 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 5.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.7 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.3 | 1.0 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.3 | 1.3 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.2 | 0.7 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 1.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.2 | 1.9 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.2 | 2.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 2.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 1.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.5 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 1.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.9 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 1.1 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 0.5 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 0.6 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 1.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.3 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 1.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.6 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 1.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 1.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 3.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.3 | GO:0008665 | tRNA 2'-phosphotransferase activity(GO:0000215) 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 0.4 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 1.0 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 0.9 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 0.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 2.8 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.1 | 0.5 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 1.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 2.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.0 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 2.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 2.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.3 | GO:0008009 | chemokine activity(GO:0008009) chemokine receptor binding(GO:0042379) |
| 0.0 | 0.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.9 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.1 | GO:0070042 | rRNA (uridine-N3-)-methyltransferase activity(GO:0070042) |
| 0.0 | 3.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 1.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.9 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 17.2 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 2.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.8 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 3.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.0 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.2 | 1.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 0.6 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 2.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 2.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 2.2 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.4 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.4 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |