Project
DANIO-CODE
Navigation
Downloads
Results for LHX6
Z-value: 0.98
Transcription factors associated with LHX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LHX6
|
ENSDARG00000052165 | si_ch211-236k19.2 |
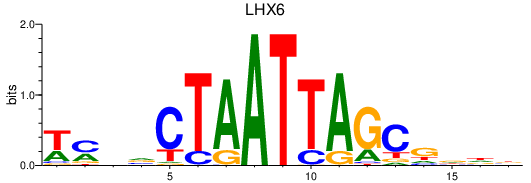
Activity profile of LHX6 motif
Sorted Z-values of LHX6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of LHX6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_18564182 | 4.15 |
ENSDART00000176809
|
sfxn3
|
sideroflexin 3 |
| chr17_-_45021393 | 3.19 |
|
|
|
| chr12_+_47448318 | 2.45 |
ENSDART00000152857
|
fmn2b
|
formin 2b |
| chr10_-_34971985 | 2.38 |
ENSDART00000141201
|
ccna1
|
cyclin A1 |
| chr22_+_37688839 | 2.30 |
ENSDART00000174664
ENSDART00000178200 |
FO704589.1
|
ENSDARG00000108910 |
| chr24_+_19270877 | 2.29 |
|
|
|
| chr5_+_37303599 | 1.99 |
ENSDART00000097754
ENSDART00000162470 |
tmprss4b
|
transmembrane protease, serine 4b |
| chr16_+_47283253 | 1.99 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr8_-_23759076 | 1.96 |
ENSDART00000145894
|
zgc:195245
|
zgc:195245 |
| chr2_+_3115593 | 1.95 |
ENSDART00000160715
|
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr3_+_17783319 | 1.80 |
ENSDART00000104299
|
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr9_+_18821640 | 1.79 |
ENSDART00000006514
|
gtf2f2b
|
general transcription factor IIF, polypeptide 2b |
| chr3_+_17562348 | 1.72 |
|
|
|
| chr14_-_14353451 | 1.59 |
ENSDART00000170355
ENSDART00000159888 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr12_-_33256671 | 1.57 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr11_-_44756789 | 1.53 |
ENSDART00000161712
|
syngr2b
|
synaptogyrin 2b |
| chr2_-_57020663 | 1.51 |
|
|
|
| chr4_+_9466175 | 1.51 |
ENSDART00000012659
|
zgc:55888
|
zgc:55888 |
| chr3_-_5157662 | 1.51 |
ENSDART00000169609
|
tefb
|
thyrotrophic embryonic factor b |
| chr4_+_4825461 | 1.49 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr3_-_20912934 | 1.47 |
ENSDART00000156275
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr22_-_26254136 | 1.44 |
ENSDART00000060978
|
wdr83
|
WD repeat domain containing 83 |
| chr6_+_28218420 | 1.42 |
ENSDART00000171216
ENSDART00000171377 |
si:ch73-14h10.2
|
si:ch73-14h10.2 |
| chr13_-_37530793 | 1.41 |
ENSDART00000141295
|
si:dkey-188i13.11
|
si:dkey-188i13.11 |
| chr18_-_43890836 | 1.38 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr11_-_15886860 | 1.34 |
ENSDART00000170731
|
zgc:173544
|
zgc:173544 |
| chr12_-_33256754 | 1.33 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr6_-_46740359 | 1.32 |
ENSDART00000011970
|
zgc:66479
|
zgc:66479 |
| chr16_+_47283374 | 1.29 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| KN150663v1_-_3381 | 1.29 |
|
|
|
| chr12_-_22116694 | 1.25 |
ENSDART00000038310
ENSDART00000132731 |
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr25_-_13394261 | 1.25 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr19_-_4876806 | 1.24 |
ENSDART00000141336
ENSDART00000110551 |
st3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr1_+_29919378 | 1.23 |
ENSDART00000127670
|
bora
|
bora, aurora kinase A activator |
| chr7_+_6918301 | 1.21 |
ENSDART00000109485
|
gal3st3
|
galactose-3-O-sulfotransferase 3 |
| chr6_-_8125341 | 1.20 |
ENSDART00000139161
ENSDART00000140021 |
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr7_+_69290790 | 1.19 |
ENSDART00000109507
|
RAP1GDS1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr17_+_7377230 | 1.17 |
ENSDART00000157123
|
shprh
|
SNF2 histone linker PHD RING helicase |
| chr23_+_43868027 | 1.17 |
ENSDART00000112598
ENSDART00000169576 |
otud4
|
OTU deubiquitinase 4 |
| chr22_+_4035577 | 1.15 |
ENSDART00000170620
|
ctxn1
|
cortexin 1 |
| chr12_-_1435708 | 1.12 |
|
|
|
| chr18_-_14306241 | 1.11 |
ENSDART00000166643
|
mlycd
|
malonyl-CoA decarboxylase |
| chr17_+_52526741 | 1.10 |
ENSDART00000109891
|
angel1
|
angel homolog 1 (Drosophila) |
| chr5_-_39692560 | 1.09 |
ENSDART00000097525
|
snx18a
|
sorting nexin 18a |
| chr12_-_33257026 | 1.09 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr16_+_42567707 | 1.09 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr19_-_12728868 | 1.08 |
ENSDART00000103692
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr13_+_12629211 | 1.08 |
ENSDART00000015127
|
zgc:100846
|
zgc:100846 |
| chr8_+_11287550 | 1.07 |
ENSDART00000115057
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr10_-_34971926 | 1.07 |
ENSDART00000141201
|
ccna1
|
cyclin A1 |
| chr3_+_48607256 | 1.04 |
ENSDART00000168449
|
raver1
|
ribonucleoprotein, PTB-binding 1 |
| chr2_+_6341404 | 1.04 |
ENSDART00000076700
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr23_-_33692244 | 1.03 |
|
|
|
| chr10_-_32550351 | 1.02 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr2_-_3115484 | 1.02 |
|
|
|
| chr1_+_13244109 | 1.01 |
ENSDART00000157563
|
noctb
|
nocturnin b |
| chr12_-_33256934 | 1.01 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr12_+_33257120 | 0.99 |
|
|
|
| chr13_-_37530730 | 0.97 |
ENSDART00000141295
|
si:dkey-188i13.11
|
si:dkey-188i13.11 |
| chr20_-_37910887 | 0.97 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr25_-_1243081 | 0.95 |
ENSDART00000156062
|
calml4b
|
calmodulin-like 4b |
| chr9_+_45193290 | 0.94 |
ENSDART00000176175
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr13_+_8622628 | 0.92 |
ENSDART00000100241
|
haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr14_+_7596207 | 0.92 |
ENSDART00000113299
|
zgc:110843
|
zgc:110843 |
| chr6_+_40925259 | 0.92 |
ENSDART00000002728
ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| KN149790v1_-_67669 | 0.92 |
ENSDART00000163561
|
ENSDARG00000099322
|
ENSDARG00000099322 |
| chr12_-_33256599 | 0.91 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr3_-_30754236 | 0.90 |
ENSDART00000109104
|
suv420h2
|
suppressor of variegation 4-20 homolog 2 (Drosophila) |
| chr24_+_17115897 | 0.90 |
ENSDART00000129554
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr1_+_13244145 | 0.89 |
ENSDART00000157563
|
noctb
|
nocturnin b |
| chr7_+_22042296 | 0.89 |
ENSDART00000123457
|
TMEM102
|
transmembrane protein 102 |
| chr22_-_21873054 | 0.88 |
|
|
|
| chr17_+_22291644 | 0.85 |
ENSDART00000151929
ENSDART00000089919 ENSDART00000000804 |
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr14_-_14353487 | 0.85 |
ENSDART00000172241
|
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr7_+_6918384 | 0.84 |
ENSDART00000109485
|
gal3st3
|
galactose-3-O-sulfotransferase 3 |
| chr3_-_7275821 | 0.84 |
ENSDART00000132197
|
BX005085.4
|
ENSDARG00000102658 |
| chr3_+_26682186 | 0.83 |
ENSDART00000055537
|
socs1a
|
suppressor of cytokine signaling 1a |
| chr9_+_39309903 | 0.83 |
|
|
|
| chr21_-_39521698 | 0.83 |
ENSDART00000020174
|
dynll2b
|
dynein, light chain, LC8-type 2b |
| chr8_-_14042703 | 0.81 |
ENSDART00000042867
|
dedd
|
death effector domain containing |
| chr5_+_60823823 | 0.80 |
ENSDART00000023676
|
bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr4_+_9466147 | 0.80 |
ENSDART00000012659
|
zgc:55888
|
zgc:55888 |
| chr2_+_3115541 | 0.80 |
ENSDART00000160715
|
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr12_+_23939869 | 0.80 |
ENSDART00000021298
|
asb3
|
ankyrin repeat and SOCS box containing 3 |
| chr1_-_55195566 | 0.79 |
ENSDART00000019936
|
prkacab
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate b |
| chr16_-_28723759 | 0.79 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr15_+_25503446 | 0.79 |
ENSDART00000154164
|
aifm4
|
apoptosis-inducing factor, mitochondrion-associated, 4 |
| chr24_-_17261929 | 0.75 |
ENSDART00000153858
|
BX005392.2
|
ENSDARG00000096996 |
| chr7_+_72925910 | 0.75 |
ENSDART00000175267
ENSDART00000179638 |
CABZ01067170.1
|
ENSDARG00000106453 |
| chr18_-_26995562 | 0.74 |
ENSDART00000147823
|
rcn2
|
reticulocalbin 2 |
| chr25_+_3169073 | 0.74 |
|
|
|
| chr5_+_12894381 | 0.73 |
ENSDART00000051669
|
tctn2
|
tectonic family member 2 |
| chr2_+_3115688 | 0.73 |
ENSDART00000160715
|
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr23_-_31119917 | 0.73 |
|
|
|
| chr7_+_20092235 | 0.72 |
ENSDART00000139274
|
ponzr1
|
plac8 onzin related protein 1 |
| chr9_+_33310981 | 0.72 |
|
|
|
| chr12_+_22459218 | 0.72 |
ENSDART00000171725
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr22_-_22234380 | 0.72 |
ENSDART00000020937
|
hdgfrp2
|
hepatoma-derived growth factor-related protein 2 |
| chr5_+_25133592 | 0.70 |
ENSDART00000098467
|
abhd17b
|
abhydrolase domain containing 17B |
| chr7_-_40359613 | 0.69 |
ENSDART00000173670
|
ube3c
|
ubiquitin protein ligase E3C |
| chr7_+_20091996 | 0.69 |
ENSDART00000138786
|
ponzr1
|
plac8 onzin related protein 1 |
| chr25_-_10694465 | 0.69 |
ENSDART00000127054
|
BX572619.1
|
ENSDARG00000089303 |
| chr5_+_60782855 | 0.67 |
ENSDART00000074073
|
alkbh4
|
alkB homolog 4, lysine demthylase |
| chr11_+_42310157 | 0.67 |
ENSDART00000085868
|
appl1
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 |
| chr12_+_5013049 | 0.66 |
ENSDART00000161548
|
kif22
|
kinesin family member 22 |
| chr5_+_55763413 | 0.65 |
|
|
|
| chr23_+_37124543 | 0.64 |
ENSDART00000155706
|
CR762407.2
|
ENSDARG00000097111 |
| chr22_-_20378023 | 0.64 |
ENSDART00000010048
|
map2k2a
|
mitogen-activated protein kinase kinase 2a |
| chr13_-_24695520 | 0.63 |
ENSDART00000142745
|
slka
|
STE20-like kinase a |
| chr21_+_4091665 | 0.63 |
ENSDART00000148138
|
lrrc8aa
|
leucine rich repeat containing 8 family, member Aa |
| chr4_-_75683584 | 0.63 |
ENSDART00000075770
|
zgc:162948
|
zgc:162948 |
| chr10_-_25808063 | 0.62 |
ENSDART00000134176
|
postna
|
periostin, osteoblast specific factor a |
| chr12_+_22459177 | 0.61 |
ENSDART00000171725
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr5_+_60823598 | 0.61 |
ENSDART00000023676
|
bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr25_+_20596490 | 0.61 |
ENSDART00000073648
|
kxd1
|
KxDL motif containing 1 |
| chr12_-_1435616 | 0.60 |
|
|
|
| chr3_-_26073798 | 0.59 |
ENSDART00000169123
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr6_-_8125287 | 0.59 |
ENSDART00000139161
ENSDART00000140021 |
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr3_-_27784565 | 0.59 |
ENSDART00000115370
|
mettl22
|
methyltransferase like 22 |
| chr5_+_6005049 | 0.59 |
|
|
|
| chr18_+_5707331 | 0.58 |
|
|
|
| chr23_+_36494483 | 0.57 |
ENSDART00000042701
|
pip4k2ca
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma a |
| chr17_+_49540985 | 0.57 |
|
|
|
| chr12_+_3632548 | 0.56 |
|
|
|
| chr7_+_15059782 | 0.56 |
ENSDART00000165683
|
mespba
|
mesoderm posterior ba |
| chr21_-_39132507 | 0.56 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr10_+_16078021 | 0.55 |
ENSDART00000065037
ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr3_+_12966604 | 0.55 |
ENSDART00000172460
|
PRKAR1B (1 of many)
|
protein kinase cAMP-dependent type I regulatory subunit beta |
| chr19_+_47918720 | 0.54 |
ENSDART00000157886
|
zgc:114119
|
zgc:114119 |
| chr10_+_27028355 | 0.54 |
ENSDART00000064111
|
faub
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed b |
| chr13_-_36537892 | 0.53 |
ENSDART00000085319
|
sos2
|
son of sevenless homolog 2 (Drosophila) |
| chr24_-_9838947 | 0.53 |
ENSDART00000138576
|
zgc:171977
|
zgc:171977 |
| chr20_+_24047842 | 0.53 |
ENSDART00000128616
ENSDART00000144195 |
casp8ap2
|
caspase 8 associated protein 2 |
| chr4_+_4825409 | 0.52 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr10_+_34057791 | 0.51 |
ENSDART00000149934
|
kl
|
klotho |
| chr22_-_58352 | 0.51 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr4_+_76572863 | 0.50 |
ENSDART00000156692
|
arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr2_+_42923652 | 0.50 |
ENSDART00000168318
|
BX005083.1
|
ENSDARG00000097562 |
| chr12_+_23939934 | 0.49 |
ENSDART00000021298
|
asb3
|
ankyrin repeat and SOCS box containing 3 |
| chr1_+_29919729 | 0.49 |
ENSDART00000127670
|
bora
|
bora, aurora kinase A activator |
| chr4_+_4825628 | 0.48 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr25_+_3634007 | 0.48 |
ENSDART00000055845
|
thoc5
|
THO complex 5 |
| chr5_+_37303707 | 0.48 |
ENSDART00000097754
ENSDART00000162470 |
tmprss4b
|
transmembrane protease, serine 4b |
| chr12_+_27498216 | 0.48 |
ENSDART00000066288
|
spata20
|
spermatogenesis associated 20 |
| chr1_+_29919605 | 0.46 |
ENSDART00000127670
|
bora
|
bora, aurora kinase A activator |
| chr2_+_10209233 | 0.46 |
ENSDART00000160304
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr1_-_54505112 | 0.45 |
ENSDART00000100619
ENSDART00000163796 |
LUC7L2
|
zgc:158803 |
| chr3_+_32802899 | 0.45 |
|
|
|
| chr5_-_39692410 | 0.44 |
ENSDART00000097525
|
snx18a
|
sorting nexin 18a |
| chr12_+_37039816 | 0.44 |
ENSDART00000162922
|
CR456627.1
|
ENSDARG00000104543 |
| chr3_+_19829535 | 0.44 |
|
|
|
| chr24_-_24578984 | 0.43 |
ENSDART00000012399
|
armc1
|
armadillo repeat containing 1 |
| chr22_+_37696341 | 0.43 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr6_+_50452131 | 0.43 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr9_-_12623750 | 0.43 |
ENSDART00000142024
|
tra2b
|
transformer 2 beta homolog |
| chr13_+_12629243 | 0.43 |
ENSDART00000015127
|
zgc:100846
|
zgc:100846 |
| chr6_+_58838038 | 0.42 |
ENSDART00000103144
ENSDART00000042595 |
dctn2
|
dynactin 2 (p50) |
| chr1_+_18740188 | 0.42 |
ENSDART00000144403
|
clockb
|
clock circadian regulator b |
| chr17_+_8054784 | 0.42 |
ENSDART00000133301
|
slc18b1
|
solute carrier family 18, subfamily B, member 1 |
| chr7_+_72030256 | 0.42 |
ENSDART00000172021
|
tollip
|
toll interacting protein |
| chr12_+_35849423 | 0.41 |
|
|
|
| chr14_+_21522672 | 0.41 |
ENSDART00000122739
|
ctbp1
|
C-terminal binding protein 1 |
| chr19_+_12487051 | 0.41 |
ENSDART00000052238
ENSDART00000013865 |
seh1l
|
SEH1-like (S. cerevisiae) |
| chr2_-_36845638 | 0.41 |
|
|
|
| chr24_-_36227144 | 0.41 |
|
|
|
| chr3_-_30754325 | 0.41 |
ENSDART00000109104
|
suv420h2
|
suppressor of variegation 4-20 homolog 2 (Drosophila) |
| chr22_-_26254217 | 0.40 |
ENSDART00000060978
|
wdr83
|
WD repeat domain containing 83 |
| chr22_-_22234212 | 0.40 |
ENSDART00000113824
ENSDART00000169594 ENSDART00000158261 |
hdgfrp2
|
hepatoma-derived growth factor-related protein 2 |
| chr21_-_18238558 | 0.40 |
ENSDART00000126672
|
brd3a
|
bromodomain containing 3a |
| chr8_+_8908955 | 0.40 |
ENSDART00000131215
|
slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member 2 |
| chr21_-_13565401 | 0.40 |
ENSDART00000151547
|
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr24_+_16402587 | 0.39 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr3_+_32234155 | 0.39 |
ENSDART00000155967
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr18_-_21961658 | 0.39 |
ENSDART00000043452
|
tsnaxip1
|
translin-associated factor X interacting protein 1 |
| chr6_-_1685041 | 0.39 |
|
|
|
| chr16_+_42567668 | 0.39 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr21_-_151444 | 0.38 |
|
|
|
| chr23_+_38313746 | 0.37 |
ENSDART00000129593
|
znf217
|
zinc finger protein 217 |
| chr1_-_22577855 | 0.37 |
ENSDART00000175685
|
rfc1
|
replication factor C (activator 1) 1 |
| chr21_-_16442202 | 0.37 |
|
|
|
| chr2_+_6341345 | 0.36 |
ENSDART00000058256
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr24_+_37752868 | 0.36 |
ENSDART00000158181
|
wdr24
|
WD repeat domain 24 |
| chr8_-_16428630 | 0.36 |
ENSDART00000098691
|
rnf11b
|
ring finger protein 11b |
| chr16_-_54679352 | 0.35 |
ENSDART00000083713
|
clk2b
|
CDC-like kinase 2b |
| chr22_-_21872864 | 0.34 |
ENSDART00000158501
|
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr3_+_33788977 | 0.34 |
ENSDART00000055248
|
alkbh7
|
alkB homolog 7 |
| chr13_-_36538099 | 0.33 |
|
|
|
| chr25_-_2570515 | 0.33 |
ENSDART00000143721
|
adpgk
|
ADP-dependent glucokinase |
| chr6_+_18522756 | 0.32 |
ENSDART00000156169
|
ube2o
|
ubiquitin-conjugating enzyme E2O |
| chr16_-_28723722 | 0.32 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr19_+_46371192 | 0.32 |
ENSDART00000169710
|
utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr14_-_11714513 | 0.31 |
ENSDART00000106654
|
znf711
|
zinc finger protein 711 |
| chr19_+_9066225 | 0.30 |
ENSDART00000018973
|
scamp3
|
secretory carrier membrane protein 3 |
| chr1_+_518777 | 0.30 |
ENSDART00000109083
|
txnl4b
|
thioredoxin-like 4B |
| chr5_+_32751876 | 0.30 |
|
|
|
| chr24_+_26241878 | 0.29 |
|
|
|
| chr2_-_2252088 | 0.29 |
ENSDART00000164674
|
ss18
|
synovial sarcoma translocation, chromosome 18 (H. sapiens) |
| chr5_-_21548930 | 0.29 |
ENSDART00000002938
|
mtmr8
|
myotubularin related protein 8 |
| chr12_+_22459371 | 0.28 |
ENSDART00000171725
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr13_-_48884956 | 0.28 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.4 | 1.1 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.3 | 0.9 | GO:0043420 | anthranilate metabolic process(GO:0043420) |
| 0.3 | 1.2 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.3 | 0.8 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.3 | 1.8 | GO:0045453 | bone resorption(GO:0045453) |
| 0.3 | 1.5 | GO:0036268 | swimming(GO:0036268) |
| 0.2 | 3.5 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.2 | 3.0 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.2 | 0.6 | GO:1902635 | 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate metabolic process(GO:1902633) 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process(GO:1902635) |
| 0.2 | 1.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 0.5 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.2 | 0.8 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 1.8 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.2 | 1.4 | GO:0072088 | renal tubule morphogenesis(GO:0061333) nephron tubule morphogenesis(GO:0072078) nephron epithelium morphogenesis(GO:0072088) |
| 0.1 | 0.6 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.7 | GO:0072584 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 2.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 5.9 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.1 | 2.3 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 0.7 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 0.9 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.1 | 1.3 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 0.9 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 1.4 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.4 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 | 1.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 1.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 2.6 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 3.7 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 4.2 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.1 | 0.7 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 0.1 | 0.7 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.5 | GO:0071482 | cellular response to light stimulus(GO:0071482) |
| 0.1 | 0.3 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.2 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.6 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 1.3 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 1.4 | GO:0010906 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.7 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.4 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.3 | GO:0097300 | necrotic cell death(GO:0070265) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.1 | GO:0035264 | multicellular organism growth(GO:0035264) |
| 0.0 | 2.1 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.4 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 1.2 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.5 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 1.2 | GO:0072329 | monocarboxylic acid catabolic process(GO:0072329) |
| 0.0 | 1.1 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 0.5 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.4 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.3 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 1.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.2 | GO:0006415 | translational termination(GO:0006415) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.4 | 1.8 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.4 | 1.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.3 | 3.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.2 | 1.1 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.4 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 1.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 4.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.7 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.3 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 0.9 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 0.5 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 3.2 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.4 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.7 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 1.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.9 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.6 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.0 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.2 | GO:0000792 | heterochromatin(GO:0000792) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.6 | 4.2 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.5 | 2.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.4 | 1.2 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.4 | 1.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.3 | 2.4 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.3 | 1.4 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 2.0 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 0.6 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 1.1 | GO:0015562 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) efflux transmembrane transporter activity(GO:0015562) |
| 0.2 | 1.4 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 4.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 1.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 1.8 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.1 | 0.9 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 2.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 1.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.5 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.9 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.8 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.9 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 0.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.1 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 3.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.0 | 0.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 1.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.2 | GO:0001047 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.0 | 3.0 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 2.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 1.8 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 1.1 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.7 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.6 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 4.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.8 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 1.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 3.7 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 0.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |