Project
DANIO-CODE
Navigation
Downloads
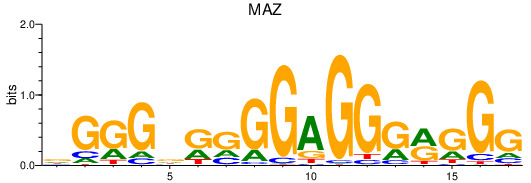
Results for MAZ
Z-value: 6.68
Transcription factors associated with MAZ
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MAZ
|
ENSDARG00000063555 | si_ch211-166g5.4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ENSDARG00000063555 | dr10_dc_chr12_-_3741925_3741966 | 0.90 | 2.0e-06 | Click! |
Activity profile of MAZ motif
Sorted Z-values of MAZ motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MAZ
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_44330405 | 25.35 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr3_-_32202533 | 21.91 |
ENSDART00000155757
|
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr7_-_57796486 | 16.40 |
ENSDART00000043984
|
ank2b
|
ankyrin 2b, neuronal |
| chr7_-_60525749 | 14.89 |
ENSDART00000136999
|
pcxb
|
pyruvate carboxylase b |
| chr25_+_31769985 | 14.36 |
|
|
|
| chr19_+_31046291 | 14.03 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr7_+_25758469 | 13.95 |
ENSDART00000101126
|
alox12
|
arachidonate 12-lipoxygenase |
| chr7_+_39130411 | 13.50 |
ENSDART00000144750
|
tnni2b.1
|
troponin I type 2b (skeletal, fast), tandem duplicate 1 |
| chr12_-_26760324 | 13.40 |
ENSDART00000047724
|
zeb1b
|
zinc finger E-box binding homeobox 1b |
| chr8_-_11191783 | 13.27 |
ENSDART00000131171
|
unc45b
|
unc-45 myosin chaperone B |
| chr19_-_19290260 | 13.25 |
|
|
|
| chr10_+_10252074 | 13.24 |
ENSDART00000144214
|
sh2d3ca
|
SH2 domain containing 3Ca |
| chr23_-_35657313 | 12.33 |
ENSDART00000043429
|
jph2
|
junctophilin 2 |
| chr14_+_30390575 | 12.18 |
ENSDART00000087884
|
ccdc85b
|
coiled-coil domain containing 85B |
| chr19_+_42491631 | 12.00 |
ENSDART00000150949
|
nfyc
|
nuclear transcription factor Y, gamma |
| chr7_+_44373815 | 11.82 |
ENSDART00000170721
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr24_+_13780640 | 11.60 |
ENSDART00000136443
ENSDART00000081595 ENSDART00000012253 |
eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr23_-_27226280 | 11.47 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr23_-_15807415 | 11.47 |
ENSDART00000178557
|
BX005011.3
|
ENSDARG00000108083 |
| chr8_-_9081109 | 11.45 |
ENSDART00000176850
|
slc6a8
|
solute carrier family 6 (neurotransmitter transporter), member 8 |
| chr7_+_34482282 | 11.40 |
|
|
|
| chr13_-_36996246 | 11.36 |
ENSDART00000133242
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr20_+_14979154 | 11.31 |
ENSDART00000118157
|
dre-mir-199-1
|
dre-mir-199-1 |
| chr24_+_26944182 | 11.30 |
ENSDART00000089541
|
dip2ca
|
disco-interacting protein 2 homolog Ca |
| chr3_+_26896869 | 11.29 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr19_-_46396376 | 11.18 |
|
|
|
| chr4_-_17066886 | 10.98 |
ENSDART00000134595
|
sox5
|
SRY (sex determining region Y)-box 5 |
| chr19_-_6384776 | 10.93 |
|
|
|
| chr16_+_23516127 | 10.60 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr3_+_53518347 | 10.46 |
ENSDART00000170461
|
col5a3a
|
collagen, type V, alpha 3a |
| chr14_-_9216303 | 10.44 |
ENSDART00000054689
|
atoh8
|
atonal bHLH transcription factor 8 |
| chr20_-_21773202 | 10.36 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr14_-_25279835 | 10.28 |
ENSDART00000163669
|
cplx2
|
complexin 2 |
| chr11_-_29316162 | 10.27 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr11_-_32461160 | 10.26 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr10_+_31358236 | 10.25 |
ENSDART00000145562
|
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr24_-_4941954 | 10.23 |
ENSDART00000127597
|
zic1
|
zic family member 1 (odd-paired homolog, Drosophila) |
| chr21_-_11554253 | 10.23 |
ENSDART00000133443
|
cast
|
calpastatin |
| chr1_-_37990863 | 10.17 |
ENSDART00000132402
|
gpm6ab
|
glycoprotein M6Ab |
| chr6_+_37881015 | 10.14 |
ENSDART00000087311
|
oca2
|
oculocutaneous albinism II |
| chr3_-_60929921 | 9.97 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr7_+_21455138 | 9.97 |
ENSDART00000173992
|
kdm6ba
|
lysine (K)-specific demethylase 6B, a |
| chr19_-_2399429 | 9.88 |
ENSDART00000043595
|
twist1a
|
twist family bHLH transcription factor 1a |
| chr2_-_4277516 | 9.86 |
|
|
|
| chr20_+_22321303 | 9.86 |
ENSDART00000049204
|
kdr
|
kinase insert domain receptor (a type III receptor tyrosine kinase) |
| chr4_-_12863235 | 9.79 |
ENSDART00000080536
|
hmga2
|
high mobility group AT-hook 2 |
| chr17_+_48999061 | 9.78 |
ENSDART00000177390
|
tiam2a
|
T-cell lymphoma invasion and metastasis 2a |
| KN149946v1_-_12861 | 9.78 |
ENSDART00000163016
|
CABZ01048402.2
|
ENSDARG00000103875 |
| chr11_-_25592102 | 9.77 |
ENSDART00000103549
|
skib
|
v-ski avian sarcoma viral oncogene homolog b |
| chr18_-_45750 | 9.66 |
ENSDART00000148821
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr21_-_43020159 | 9.61 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr5_-_13826859 | 9.59 |
ENSDART00000137355
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr19_-_5338129 | 9.58 |
ENSDART00000081951
|
stx1b
|
syntaxin 1B |
| chr4_-_9172622 | 9.53 |
ENSDART00000042963
|
chst11
|
carbohydrate (chondroitin 4) sulfotransferase 11 |
| chr10_+_10252239 | 9.44 |
ENSDART00000144214
|
sh2d3ca
|
SH2 domain containing 3Ca |
| chr7_+_20699191 | 9.41 |
ENSDART00000062003
|
efnb3b
|
ephrin-B3b |
| chr7_+_35769973 | 9.38 |
ENSDART00000168658
|
irx3a
|
iroquois homeobox 3a |
| chr2_-_39710140 | 9.34 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr8_+_28493742 | 9.26 |
ENSDART00000053782
|
scrt2
|
scratch family zinc finger 2 |
| chr18_-_39602605 | 9.20 |
ENSDART00000125116
|
tnfaip8l3
|
tumor necrosis factor, alpha-induced protein 8-like 3 |
| chr20_-_54192057 | 9.19 |
ENSDART00000023550
|
hsp90aa1.2
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 2 |
| chr19_+_4996328 | 9.07 |
ENSDART00000165082
|
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr18_-_11216447 | 9.02 |
ENSDART00000040500
|
tspan9a
|
tetraspanin 9a |
| chr4_+_11312432 | 8.88 |
ENSDART00000051792
|
sema3aa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Aa |
| chr20_-_29580624 | 8.83 |
ENSDART00000062370
|
actc1a
|
actin, alpha, cardiac muscle 1a |
| chr9_+_14180954 | 8.81 |
ENSDART00000148055
|
si:ch211-67e16.11
|
si:ch211-67e16.11 |
| chr8_-_52952226 | 8.79 |
ENSDART00000168185
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr7_-_23965038 | 8.70 |
ENSDART00000010124
|
slc7a8a
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8a |
| chr3_-_38550961 | 8.65 |
ENSDART00000155042
|
mpp3a
|
membrane protein, palmitoylated 3a (MAGUK p55 subfamily member 3) |
| chr5_+_4372660 | 8.64 |
ENSDART00000067599
|
angptl2a
|
angiopoietin-like 2a |
| chr14_+_21531709 | 8.64 |
ENSDART00000144367
|
ctbp1
|
C-terminal binding protein 1 |
| chr25_-_2485276 | 8.63 |
ENSDART00000154889
ENSDART00000155027 |
BX957346.1
|
ENSDARG00000096850 |
| chr7_-_71958596 | 8.61 |
|
|
|
| chr9_+_54688205 | 8.53 |
ENSDART00000067544
|
egfl6
|
EGF-like-domain, multiple 6 |
| chr7_-_41188744 | 8.50 |
ENSDART00000174087
|
smarcd3b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3b |
| chr3_+_32293441 | 8.45 |
ENSDART00000156464
|
prr12b
|
proline rich 12b |
| chr17_+_52736192 | 8.45 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr23_-_31502149 | 8.38 |
ENSDART00000053545
|
zgc:153284
|
zgc:153284 |
| chr7_-_52283383 | 8.38 |
ENSDART00000165649
|
tcf12
|
transcription factor 12 |
| chr19_-_5142310 | 8.35 |
ENSDART00000130062
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr2_+_23038657 | 8.27 |
ENSDART00000089012
|
kif1ab
|
kinesin family member 1Ab |
| chr8_+_39964695 | 8.22 |
ENSDART00000073782
|
ggt5a
|
gamma-glutamyltransferase 5a |
| chr17_+_22359660 | 8.21 |
ENSDART00000162670
|
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr2_+_43619752 | 8.14 |
ENSDART00000142078
ENSDART00000098265 ENSDART00000098267 |
nrp1b
|
neuropilin 1b |
| chr20_+_30917073 | 8.13 |
|
|
|
| chr23_-_27226387 | 8.11 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr13_-_49826330 | 8.10 |
ENSDART00000099475
|
nid1a
|
nidogen 1a |
| chr18_+_44656323 | 8.10 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr16_-_51390400 | 8.03 |
ENSDART00000148894
|
serpinb14
|
serpin peptidase inhibitor, clade B (ovalbumin), member 14 |
| chr14_+_23986703 | 7.96 |
ENSDART00000079164
|
klhl3
|
kelch-like family member 3 |
| chr19_+_9113356 | 7.93 |
ENSDART00000127755
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr11_-_25592017 | 7.91 |
ENSDART00000103549
|
skib
|
v-ski avian sarcoma viral oncogene homolog b |
| chr20_+_26408687 | 7.89 |
|
|
|
| chr13_-_508202 | 7.86 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr19_+_42492029 | 7.85 |
ENSDART00000150949
|
nfyc
|
nuclear transcription factor Y, gamma |
| chr18_+_12207888 | 7.82 |
|
|
|
| chr22_-_95158 | 7.77 |
|
|
|
| chr19_-_47994946 | 7.76 |
ENSDART00000114549
|
CU695215.1
|
ENSDARG00000076126 |
| chr16_+_23172295 | 7.71 |
ENSDART00000167518
ENSDART00000161087 |
efna3b
|
ephrin-A3b |
| chr16_-_37461878 | 7.69 |
ENSDART00000142916
|
si:ch211-208k15.1
|
si:ch211-208k15.1 |
| chr3_-_24925189 | 7.68 |
ENSDART00000156459
|
ep300b
|
E1A binding protein p300 b |
| chr20_-_26521700 | 7.65 |
ENSDART00000110883
|
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr24_+_28364360 | 7.64 |
ENSDART00000018037
|
arhgap29a
|
Rho GTPase activating protein 29a |
| chr7_-_7164950 | 7.63 |
ENSDART00000012637
|
zgc:101810
|
zgc:101810 |
| chr6_+_2849460 | 7.60 |
|
|
|
| chr19_-_2011177 | 7.55 |
ENSDART00000108784
|
mturn
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr1_-_18709814 | 7.55 |
ENSDART00000145224
|
rbm47
|
RNA binding motif protein 47 |
| chr10_-_31619761 | 7.43 |
ENSDART00000023575
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr4_-_12862869 | 7.36 |
ENSDART00000080536
|
hmga2
|
high mobility group AT-hook 2 |
| chr5_-_47520995 | 7.25 |
ENSDART00000144252
|
BX465834.1
|
ENSDARG00000095715 |
| chr21_+_26660833 | 7.22 |
ENSDART00000004109
|
gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr13_-_31304614 | 7.19 |
|
|
|
| chr13_-_14978365 | 7.19 |
|
|
|
| chr14_+_34146377 | 7.15 |
ENSDART00000131861
|
tmsb2
|
thymosin beta 2 |
| chr18_+_22367874 | 7.13 |
|
|
|
| KN149987v1_-_29629 | 7.08 |
ENSDART00000159555
ENSDART00000168161 |
ENSDARG00000103311
|
ENSDARG00000103311 |
| chr24_-_25283218 | 7.03 |
ENSDART00000090010
|
phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr11_-_25803101 | 7.03 |
ENSDART00000088888
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr15_-_2224784 | 6.97 |
ENSDART00000009564
|
shox2
|
short stature homeobox 2 |
| chr6_+_55355763 | 6.94 |
ENSDART00000169240
|
pltp
|
phospholipid transfer protein |
| chr2_-_32572545 | 6.92 |
ENSDART00000056641
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr25_+_30746445 | 6.91 |
ENSDART00000156916
|
lsp1
|
lymphocyte-specific protein 1 |
| chr23_-_9924987 | 6.90 |
ENSDART00000005015
|
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr12_-_10372055 | 6.87 |
ENSDART00000052001
|
eef2k
|
eukaryotic elongation factor 2 kinase |
| chr17_+_48999102 | 6.86 |
ENSDART00000177390
|
tiam2a
|
T-cell lymphoma invasion and metastasis 2a |
| chr17_-_52735615 | 6.80 |
|
|
|
| chr16_-_51390484 | 6.79 |
ENSDART00000148894
|
serpinb14
|
serpin peptidase inhibitor, clade B (ovalbumin), member 14 |
| chr8_-_23102555 | 6.79 |
|
|
|
| chr17_-_44133941 | 6.75 |
ENSDART00000126097
|
otx2
|
orthodenticle homeobox 2 |
| chr13_+_22350043 | 6.74 |
ENSDART00000136863
|
ldb3a
|
LIM domain binding 3a |
| chr5_+_37662896 | 6.73 |
ENSDART00000051231
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr1_-_50831155 | 6.73 |
ENSDART00000152719
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr16_-_12579115 | 6.69 |
ENSDART00000147483
|
ephb6
|
eph receptor B6 |
| chr3_+_17210396 | 6.65 |
ENSDART00000090676
|
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr15_-_23407388 | 6.58 |
ENSDART00000020425
|
mcamb
|
melanoma cell adhesion molecule b |
| chr22_-_21021645 | 6.57 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr9_-_49834232 | 6.56 |
|
|
|
| chr16_+_25845432 | 6.51 |
|
|
|
| chr15_+_19902697 | 6.51 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr12_+_29994735 | 6.45 |
ENSDART00000042572
ENSDART00000153025 |
ablim1b
|
actin binding LIM protein 1b |
| chr11_+_23695123 | 6.44 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr13_-_219661 | 6.40 |
ENSDART00000133731
|
si:ch1073-291c23.2
|
si:ch1073-291c23.2 |
| chr21_-_25567978 | 6.39 |
ENSDART00000133134
|
efemp2b
|
EGF containing fibulin-like extracellular matrix protein 2b |
| chr18_-_47890567 | 6.34 |
|
|
|
| chr20_+_15119519 | 6.32 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr16_+_5466342 | 6.32 |
ENSDART00000160008
|
plecb
|
plectin b |
| chr2_-_9848848 | 6.29 |
ENSDART00000056901
|
zgc:153615
|
zgc:153615 |
| chr3_-_60930025 | 6.28 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr4_-_24310846 | 6.27 |
ENSDART00000017443
|
celf2
|
cugbp, Elav-like family member 2 |
| chr13_-_18704185 | 6.27 |
ENSDART00000146795
|
lzts2a
|
leucine zipper, putative tumor suppressor 2a |
| chr25_-_4376129 | 6.26 |
|
|
|
| chr5_-_13844114 | 6.23 |
|
|
|
| chr11_-_11534819 | 6.21 |
ENSDART00000104254
|
krt15
|
keratin 15 |
| chr24_+_19374200 | 6.17 |
ENSDART00000056081
ENSDART00000027022 |
sulf1
|
sulfatase 1 |
| chr20_-_47123598 | 6.13 |
ENSDART00000100320
|
dnmt3aa
|
DNA (cytosine-5-)-methyltransferase 3 alpha a |
| chr20_+_15119754 | 6.11 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr4_+_14361566 | 6.08 |
ENSDART00000007103
|
nuak1a
|
NUAK family, SNF1-like kinase, 1a |
| chr11_-_23226859 | 6.08 |
ENSDART00000159221
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr11_-_1792616 | 6.08 |
|
|
|
| chr23_+_6298911 | 6.06 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr1_-_51862897 | 6.05 |
ENSDART00000136469
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr7_+_19300351 | 6.05 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr10_-_22834248 | 6.00 |
ENSDART00000079469
|
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr21_-_37509454 | 6.00 |
ENSDART00000175126
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr25_+_558191 | 5.93 |
ENSDART00000126863
ENSDART00000166012 |
nell2a
|
neural EGFL like 2a |
| chr20_+_316191 | 5.91 |
ENSDART00000104807
|
si:dkey-119m7.4
|
si:dkey-119m7.4 |
| chr11_-_37921642 | 5.85 |
|
|
|
| chr20_+_35377079 | 5.83 |
ENSDART00000168216
|
fam49a
|
family with sequence similarity 49, member A |
| chr5_-_13826795 | 5.82 |
ENSDART00000137355
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr19_-_27807312 | 5.82 |
|
|
|
| chr6_+_37880936 | 5.82 |
ENSDART00000087311
|
oca2
|
oculocutaneous albinism II |
| chr11_-_21143897 | 5.78 |
ENSDART00000163008
|
RASSF5
|
Ras association domain family member 5 |
| chr5_+_6054781 | 5.78 |
ENSDART00000060532
|
zgc:110796
|
zgc:110796 |
| chr1_-_28241500 | 5.75 |
ENSDART00000019770
|
gpm6ba
|
glycoprotein M6Ba |
| chr15_-_20088439 | 5.73 |
ENSDART00000161379
|
auts2b
|
autism susceptibility candidate 2b |
| chr7_-_71638662 | 5.73 |
ENSDART00000162984
|
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr20_+_6640497 | 5.73 |
ENSDART00000138361
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr15_-_17074348 | 5.67 |
ENSDART00000156768
|
hip1
|
huntingtin interacting protein 1 |
| chr23_+_34079219 | 5.67 |
ENSDART00000132668
|
si:ch211-207e14.4
|
si:ch211-207e14.4 |
| chr6_-_39278328 | 5.65 |
ENSDART00000148531
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr3_-_20888014 | 5.65 |
ENSDART00000002393
|
rundc3aa
|
RUN domain containing 3Aa |
| chr20_+_30910294 | 5.64 |
|
|
|
| chr19_+_42491275 | 5.64 |
|
|
|
| chr14_+_34207218 | 5.61 |
ENSDART00000135608
|
gabrp
|
gamma-aminobutyric acid (GABA) A receptor, pi |
| chr1_-_22144014 | 5.60 |
ENSDART00000043556
|
ldb2b
|
LIM domain binding 2b |
| chr12_+_47450923 | 5.59 |
ENSDART00000105328
|
fmn2b
|
formin 2b |
| chr5_+_8843643 | 5.57 |
ENSDART00000149417
ENSDART00000061616 |
tal2
|
T-cell acute lymphocytic leukemia 2 |
| chr14_+_45895103 | 5.57 |
|
|
|
| chr18_-_45892 | 5.57 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr24_-_34449257 | 5.56 |
ENSDART00000128690
|
agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr8_+_22910147 | 5.54 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr10_+_22759607 | 5.52 |
|
|
|
| chr14_-_6901415 | 5.48 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr18_+_21133694 | 5.47 |
ENSDART00000060015
|
chka
|
choline kinase alpha |
| chr18_+_45680282 | 5.45 |
ENSDART00000109948
|
qser1
|
glutamine and serine rich 1 |
| chr20_+_23392782 | 5.45 |
|
|
|
| chr24_-_11327087 | 5.44 |
ENSDART00000082264
|
pxdc1b
|
PX domain containing 1b |
| chr18_-_3741790 | 5.44 |
|
|
|
| chr2_-_43318809 | 5.43 |
ENSDART00000132588
|
crema
|
cAMP responsive element modulator a |
| chr20_+_33972327 | 5.42 |
ENSDART00000170930
|
rxrgb
|
retinoid X receptor, gamma b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.4 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 5.1 | 15.2 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 4.5 | 13.4 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 4.3 | 21.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 3.5 | 14.0 | GO:0019372 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 3.2 | 9.6 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 3.0 | 8.9 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) positive regulation of neuromuscular junction development(GO:1904398) |
| 2.8 | 11.3 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 2.8 | 11.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 2.7 | 16.0 | GO:0042438 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 2.6 | 10.4 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 2.5 | 7.5 | GO:2001017 | regulation of retrograde axon cargo transport(GO:2001017) |
| 2.3 | 6.9 | GO:0050810 | regulation of steroid biosynthetic process(GO:0050810) |
| 2.3 | 9.1 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 2.3 | 22.7 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 2.2 | 8.7 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 2.0 | 10.2 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 2.0 | 8.1 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 2.0 | 22.2 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 2.0 | 8.0 | GO:0070293 | renal absorption(GO:0070293) |
| 1.9 | 11.6 | GO:0016576 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 1.9 | 9.6 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 1.9 | 5.7 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 1.9 | 7.4 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 1.8 | 9.2 | GO:0046677 | response to antibiotic(GO:0046677) |
| 1.8 | 16.2 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 1.8 | 5.4 | GO:0098810 | neurotransmitter uptake(GO:0001504) regulation of neurotransmitter uptake(GO:0051580) neurotransmitter reuptake(GO:0098810) |
| 1.8 | 8.9 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 1.8 | 8.8 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 1.7 | 6.9 | GO:0034368 | protein-lipid complex remodeling(GO:0034368) plasma lipoprotein particle remodeling(GO:0034369) |
| 1.7 | 16.6 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 1.6 | 6.3 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 1.6 | 26.6 | GO:0051492 | regulation of stress fiber assembly(GO:0051492) |
| 1.5 | 10.8 | GO:0001964 | startle response(GO:0001964) |
| 1.5 | 7.5 | GO:0045639 | positive regulation of myeloid cell differentiation(GO:0045639) |
| 1.5 | 4.5 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 1.5 | 17.8 | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043516) |
| 1.4 | 11.3 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 1.4 | 9.7 | GO:0030104 | water homeostasis(GO:0030104) |
| 1.4 | 11.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 1.4 | 4.2 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 1.4 | 4.1 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 1.3 | 4.0 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 1.3 | 6.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 1.3 | 7.5 | GO:2000742 | regulation of anterior head development(GO:2000742) |
| 1.2 | 14.7 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 1.2 | 21.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 1.2 | 7.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 1.2 | 7.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 1.1 | 11.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 1.1 | 10.2 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 1.1 | 4.4 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 1.1 | 3.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 1.0 | 2.9 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.9 | 8.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.9 | 6.5 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 0.9 | 20.4 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.9 | 5.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) estrogen metabolic process(GO:0008210) |
| 0.9 | 9.6 | GO:1901797 | negative regulation of signal transduction by p53 class mediator(GO:1901797) negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.9 | 4.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.8 | 24.5 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.8 | 2.5 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.8 | 8.2 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.8 | 16.9 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.8 | 9.6 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.8 | 6.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.8 | 3.1 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.8 | 2.3 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.7 | 5.1 | GO:2001044 | regulation of integrin-mediated signaling pathway(GO:2001044) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.7 | 12.0 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.7 | 2.1 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) |
| 0.6 | 3.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.6 | 12.1 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.6 | 13.9 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.6 | 5.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.6 | 4.2 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.6 | 14.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.6 | 1.7 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.6 | 12.0 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.6 | 4.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.6 | 3.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.6 | 12.7 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.5 | 9.9 | GO:0035924 | cellular response to vascular endothelial growth factor stimulus(GO:0035924) |
| 0.5 | 3.8 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.5 | 4.2 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.5 | 11.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.5 | 7.1 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.5 | 13.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.5 | 3.4 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.5 | 4.8 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.5 | 5.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.5 | 10.3 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.5 | 1.8 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.5 | 4.5 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) dopamine transport(GO:0015872) |
| 0.4 | 3.1 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.4 | 1.3 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.4 | 8.4 | GO:0097581 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.4 | 4.9 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.4 | 10.3 | GO:0051588 | regulation of neurotransmitter secretion(GO:0046928) regulation of neurotransmitter transport(GO:0051588) |
| 0.4 | 3.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.4 | 7.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.4 | 6.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.4 | 2.7 | GO:1903038 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.4 | 10.6 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.4 | 3.5 | GO:0070654 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.4 | 5.4 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.4 | 3.4 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.4 | 3.4 | GO:0042102 | positive regulation of mononuclear cell proliferation(GO:0032946) positive regulation of T cell proliferation(GO:0042102) positive regulation of lymphocyte proliferation(GO:0050671) positive regulation of leukocyte proliferation(GO:0070665) |
| 0.4 | 14.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.4 | 3.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.4 | 1.4 | GO:0002762 | negative regulation of myeloid leukocyte differentiation(GO:0002762) |
| 0.3 | 4.2 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.3 | 1.4 | GO:0035908 | ventral aorta development(GO:0035908) pharyngeal arch artery morphogenesis(GO:0061626) |
| 0.3 | 13.3 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.3 | 4.0 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.3 | 6.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.3 | 7.8 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.3 | 7.3 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.3 | 0.9 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.3 | 1.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.3 | 10.8 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.3 | 12.0 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.3 | 2.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.3 | 14.0 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.2 | 2.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 0.7 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) |
| 0.2 | 2.9 | GO:0072576 | gland morphogenesis(GO:0022612) liver morphogenesis(GO:0072576) |
| 0.2 | 8.6 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.2 | 3.5 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.2 | 3.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.2 | 9.9 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.2 | 4.9 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.2 | 5.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.2 | 6.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.2 | 4.9 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.2 | 5.6 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.2 | 8.5 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.2 | 17.2 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.2 | 2.6 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.2 | 11.4 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.2 | 6.1 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.2 | 3.8 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.2 | 0.8 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 3.5 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.2 | 7.4 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.2 | 14.7 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.2 | 0.5 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.2 | 7.0 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.2 | 3.4 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.2 | 2.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 2.2 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.2 | 4.6 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.2 | 1.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 4.9 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 5.5 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 3.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 8.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 2.3 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 9.1 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.1 | 4.5 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.1 | 5.1 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.1 | 4.0 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.1 | 9.4 | GO:0016485 | protein processing(GO:0016485) |
| 0.1 | 1.7 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.1 | 3.4 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 5.3 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.1 | 6.7 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.1 | 4.2 | GO:0050808 | synapse organization(GO:0050808) |
| 0.1 | 3.5 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.1 | 8.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 4.5 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.1 | 11.2 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.1 | 0.4 | GO:0000305 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 1.2 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 14.0 | GO:0001525 | angiogenesis(GO:0001525) |
| 0.1 | 0.8 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 1.5 | GO:0046530 | photoreceptor cell differentiation(GO:0046530) |
| 0.0 | 1.4 | GO:0071599 | otic vesicle development(GO:0071599) |
| 0.0 | 0.4 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 0.8 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.6 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 2.2 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 3.1 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.9 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.0 | 2.1 | GO:0098609 | cell-cell adhesion(GO:0098609) |
| 0.0 | 3.0 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.2 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 2.4 | 16.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 2.2 | 11.0 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 2.0 | 19.9 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 2.0 | 7.9 | GO:0016586 | RSC complex(GO:0016586) |
| 1.5 | 21.3 | GO:0030315 | T-tubule(GO:0030315) |
| 1.4 | 22.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 1.3 | 15.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 1.1 | 5.6 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 1.1 | 4.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.9 | 9.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.8 | 11.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.8 | 13.3 | GO:0031672 | A band(GO:0031672) |
| 0.8 | 11.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.8 | 9.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.7 | 8.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.7 | 7.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.6 | 4.9 | GO:0005921 | gap junction(GO:0005921) |
| 0.6 | 13.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.6 | 4.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.6 | 6.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.6 | 4.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.5 | 10.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.5 | 5.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.5 | 21.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.5 | 9.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.5 | 22.3 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.4 | 4.9 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.4 | 2.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.4 | 7.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.4 | 12.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.3 | 10.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.3 | 16.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.3 | 3.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.3 | 17.0 | GO:0042641 | actomyosin(GO:0042641) |
| 0.3 | 8.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.3 | 9.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.3 | 3.0 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.3 | 12.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.3 | 17.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.3 | 3.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.3 | 31.1 | GO:0030424 | axon(GO:0030424) |
| 0.3 | 6.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.2 | 7.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 2.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 4.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 8.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 3.3 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 6.3 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 77.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 1.5 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 7.1 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 8.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 64.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 6.0 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 27.8 | GO:0030054 | cell junction(GO:0030054) |
| 0.1 | 3.5 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 25.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 4.5 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 22.0 | GO:0005576 | extracellular region(GO:0005576) |
| 0.1 | 2.6 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 3.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.4 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 1.0 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 2.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 3.4 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 8.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 8.8 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 8.1 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 6.7 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 2.4 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 2.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 70.8 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 1.8 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
| 0.0 | 0.8 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 14.7 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 3.1 | 15.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 3.0 | 9.1 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 2.8 | 11.4 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 2.4 | 21.9 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 2.0 | 18.0 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 1.8 | 10.8 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 1.7 | 6.9 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 1.6 | 4.9 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 1.5 | 6.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 1.4 | 5.7 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 1.4 | 5.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.4 | 11.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 1.4 | 9.6 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 1.2 | 18.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 1.2 | 3.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 1.1 | 10.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 1.1 | 5.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 1.1 | 5.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.1 | 5.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 1.1 | 5.4 | GO:2001070 | starch binding(GO:2001070) |
| 1.1 | 3.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 1.0 | 4.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 1.0 | 3.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 1.0 | 3.9 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.0 | 6.7 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.9 | 13.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.9 | 7.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.9 | 16.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.9 | 13.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.9 | 7.9 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.9 | 8.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.9 | 3.5 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.8 | 4.7 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.8 | 11.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.8 | 3.8 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.7 | 13.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.7 | 15.2 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.7 | 15.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.7 | 10.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.7 | 2.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.7 | 8.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.6 | 7.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.6 | 7.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.6 | 8.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.6 | 2.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.6 | 9.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.6 | 6.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.6 | 5.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.6 | 8.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.6 | 8.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.6 | 2.8 | GO:0015157 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.5 | 3.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.5 | 1.9 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.4 | 4.0 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.4 | 6.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.4 | 5.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.4 | 12.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.4 | 3.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.4 | 9.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.4 | 4.9 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.4 | 4.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.4 | 48.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.4 | 3.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.4 | 8.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.3 | 4.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.3 | 4.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.3 | 4.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.3 | 6.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.3 | 14.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.3 | 2.3 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.3 | 3.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 6.7 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.3 | 6.7 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.3 | 3.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.3 | 21.5 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.3 | 7.9 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.3 | 23.4 | GO:0015293 | symporter activity(GO:0015293) |
| 0.3 | 14.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 1.4 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.3 | 8.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.3 | 54.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.3 | 8.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.2 | 2.7 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.2 | 5.3 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.2 | 4.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 12.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 4.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 9.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 17.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 2.8 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.2 | 2.1 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 8.7 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.2 | 2.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 0.5 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.2 | 3.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.2 | 7.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.2 | 3.8 | GO:0016208 | AMP binding(GO:0016208) |
| 0.2 | 4.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.2 | 0.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 5.0 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 8.6 | GO:0051287 | NAD binding(GO:0051287) |
| 0.1 | 4.0 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 1.7 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 4.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 65.4 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 3.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.8 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 5.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.9 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 9.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 0.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 11.1 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.1 | 4.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 90.7 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.1 | 2.6 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 5.2 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 3.4 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 1.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 24.9 | GO:1990837 | sequence-specific double-stranded DNA binding(GO:1990837) |
| 0.1 | 9.7 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 21.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 11.8 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 3.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.7 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 1.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 1.0 | GO:0060090 | binding, bridging(GO:0060090) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.9 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 1.8 | 5.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.8 | 9.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.5 | 6.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.5 | 8.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.4 | 19.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.4 | 8.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.3 | 4.5 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.3 | 4.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.3 | 9.5 | PID ATM PATHWAY | ATM pathway |
| 0.3 | 25.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 3.9 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 7.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 4.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.2 | 4.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 2.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.2 | 16.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.2 | 9.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 4.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.7 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 3.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 3.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 9.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 1.4 | 6.9 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 1.4 | 9.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 1.3 | 13.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 1.1 | 9.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 1.1 | 11.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 1.1 | 13.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 1.0 | 6.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.9 | 2.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.8 | 9.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.6 | 5.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.6 | 7.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.5 | 11.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.5 | 3.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.4 | 3.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.4 | 2.9 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.3 | 4.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.3 | 4.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 8.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 4.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.3 | 5.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.2 | 2.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.2 | 4.6 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 4.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 22.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 1.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.6 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 4.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 3.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 2.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 3.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.9 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |