Project
DANIO-CODE
Navigation
Downloads
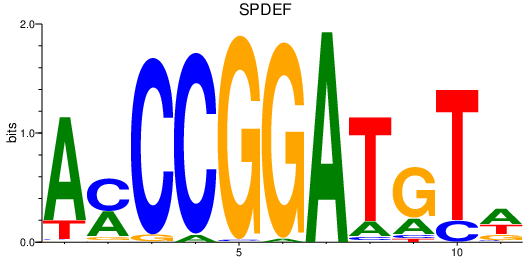
Results for SPDEF
Z-value: 0.71
Transcription factors associated with SPDEF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPDEF
|
ENSDARG00000029930 | SAM pointed domain containing ETS transcription factor |
Activity profile of SPDEF motif
Sorted Z-values of SPDEF motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SPDEF
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_22745677 | 1.61 |
ENSDART00000023721
|
nudt21
|
nudix hydrolase 21 |
| chr1_+_45907269 | 1.38 |
ENSDART00000132861
|
cab39l
|
calcium binding protein 39-like |
| chr17_-_7575628 | 1.29 |
ENSDART00000064657
|
stx11a
|
syntaxin 11a |
| chr15_+_38397772 | 1.27 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr20_-_39783396 | 1.16 |
ENSDART00000132138
ENSDART00000139252 |
BX936317.1
|
ENSDARG00000094075 |
| chr5_-_34856057 | 1.02 |
ENSDART00000051312
|
ttc33
|
tetratricopeptide repeat domain 33 |
| chr4_+_12293403 | 0.99 |
ENSDART00000061070
|
mkrn1
|
makorin, ring finger protein, 1 |
| chr7_-_27787236 | 0.97 |
ENSDART00000173842
|
RIC3
|
si:ch211-235p24.2 |
| chr8_-_14042703 | 0.89 |
ENSDART00000042867
|
dedd
|
death effector domain containing |
| chr2_+_32863386 | 0.83 |
ENSDART00000056649
|
tmem53
|
transmembrane protein 53 |
| chr5_-_26195227 | 0.81 |
ENSDART00000146124
|
si:ch211-102c2.7
|
si:ch211-102c2.7 |
| chr25_-_24150388 | 0.80 |
ENSDART00000073527
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr7_+_28906162 | 0.79 |
ENSDART00000008096
|
aph1b
|
APH1B gamma secretase subunit |
| chr2_+_52225622 | 0.79 |
ENSDART00000170353
|
ENSDARG00000103108
|
ENSDARG00000103108 |
| chr23_-_33848571 | 0.76 |
ENSDART00000027959
|
racgap1
|
Rac GTPase activating protein 1 |
| chr18_-_22745979 | 0.76 |
ENSDART00000023721
|
nudt21
|
nudix hydrolase 21 |
| chr17_-_7194149 | 0.74 |
ENSDART00000098731
|
stxbp5b
|
syntaxin binding protein 5b (tomosyn) |
| chr19_+_9536231 | 0.73 |
ENSDART00000139385
|
si:ch211-288g17.3
|
si:ch211-288g17.3 |
| chr13_-_42548129 | 0.72 |
ENSDART00000169083
|
lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr14_+_28132910 | 0.72 |
ENSDART00000006489
|
acsl4a
|
acyl-CoA synthetase long-chain family member 4a |
| chr18_-_26815192 | 0.70 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr6_-_22060441 | 0.70 |
ENSDART00000167167
|
utp6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr17_+_8596825 | 0.70 |
ENSDART00000105322
|
edrf1
|
erythroid differentiation regulatory factor 1 |
| chr13_+_31415523 | 0.69 |
ENSDART00000076527
|
ppm1aa
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Aa |
| chr15_+_19948916 | 0.69 |
ENSDART00000054416
|
dyrk1ab
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, b |
| chr22_-_10722533 | 0.69 |
ENSDART00000081191
|
sass6
|
SAS-6 centriolar assembly protein |
| chr15_-_14706032 | 0.68 |
ENSDART00000164166
|
si:dkey-260j18.2
|
si:dkey-260j18.2 |
| chr15_+_17407036 | 0.67 |
ENSDART00000018461
|
vmp1
|
vacuole membrane protein 1 |
| chr23_-_30837726 | 0.67 |
ENSDART00000075918
|
pcmtd2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr15_+_25592984 | 0.67 |
ENSDART00000123143
|
npat
|
nuclear protein, ataxia-telangiectasia locus |
| chr8_+_150970 | 0.67 |
|
|
|
| chr1_+_40894908 | 0.67 |
ENSDART00000111367
|
si:dkey-56e3.3
|
si:dkey-56e3.3 |
| chr25_-_24150237 | 0.66 |
ENSDART00000073527
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr19_+_43468369 | 0.64 |
ENSDART00000165202
|
pum1
|
pumilio RNA-binding family member 1 |
| chr20_-_49067167 | 0.63 |
ENSDART00000163071
ENSDART00000170617 |
xrn2
|
5'-3' exoribonuclease 2 |
| chr5_+_6291842 | 0.63 |
ENSDART00000148663
|
elac1
|
elaC ribonuclease Z 1 |
| chr5_-_23092592 | 0.63 |
ENSDART00000024815
|
fam76b
|
family with sequence similarity 76, member B |
| chr3_-_36298676 | 0.62 |
ENSDART00000157950
|
rogdi
|
rogdi homolog (Drosophila) |
| chr8_+_48976657 | 0.62 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr6_-_54436651 | 0.62 |
ENSDART00000154121
|
sys1
|
Sys1 golgi trafficking protein |
| chr8_+_30103357 | 0.61 |
ENSDART00000133717
|
fancc
|
Fanconi anemia, complementation group C |
| chr6_+_3174047 | 0.61 |
ENSDART00000160290
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr4_-_12979951 | 0.61 |
ENSDART00000013839
|
tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr22_-_111800 | 0.60 |
ENSDART00000163198
ENSDART00000168678 |
capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr18_+_29920669 | 0.60 |
ENSDART00000064080
|
cenpn
|
centromere protein N |
| chr1_+_45907424 | 0.59 |
ENSDART00000053240
|
cab39l
|
calcium binding protein 39-like |
| chr7_+_28905795 | 0.59 |
ENSDART00000146655
|
aph1b
|
APH1B gamma secretase subunit |
| chr5_+_24580551 | 0.59 |
ENSDART00000134526
|
paxx
|
zgc:193538 |
| KN149790v1_-_67669 | 0.57 |
ENSDART00000163561
|
ENSDARG00000099322
|
ENSDARG00000099322 |
| chr15_-_41353802 | 0.57 |
ENSDART00000085645
ENSDART00000134075 |
usp16
|
ubiquitin specific peptidase 16 |
| chr20_+_29685379 | 0.57 |
ENSDART00000178617
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr7_-_66470152 | 0.57 |
ENSDART00000021317
|
ctr9
|
CTR9 homolog, Paf1/RNA polymerase II complex component |
| chr9_+_14052296 | 0.55 |
ENSDART00000124267
|
si:ch211-67e16.4
|
si:ch211-67e16.4 |
| chr21_-_40915270 | 0.55 |
ENSDART00000008593
|
yipf5
|
Yip1 domain family, member 5 |
| KN150361v1_+_15585 | 0.55 |
|
|
|
| chr7_+_34919736 | 0.54 |
ENSDART00000110552
|
zdhhc1
|
zinc finger, DHHC-type containing 1 |
| chr24_-_32607801 | 0.53 |
ENSDART00000143781
|
EIF1B
|
eukaryotic translation initiation factor 1B |
| chr22_-_36561217 | 0.53 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr13_-_44645490 | 0.53 |
ENSDART00000099990
|
btbd9
|
BTB (POZ) domain containing 9 |
| chr13_+_31415428 | 0.53 |
ENSDART00000076527
|
ppm1aa
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Aa |
| chr22_-_4787016 | 0.53 |
ENSDART00000140313
|
tnfaip8l1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr20_+_27188382 | 0.52 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr15_+_19948869 | 0.52 |
ENSDART00000054416
|
dyrk1ab
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, b |
| chr15_+_38397715 | 0.52 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr5_-_66614185 | 0.52 |
ENSDART00000147053
|
rsrc2
|
arginine/serine-rich coiled-coil 2 |
| chr21_-_19791846 | 0.51 |
ENSDART00000037664
|
nnt
|
nicotinamide nucleotide transhydrogenase |
| chr3_-_3589775 | 0.51 |
ENSDART00000140482
|
ENSDARG00000079723
|
ENSDARG00000079723 |
| chr5_-_51363461 | 0.50 |
ENSDART00000051003
|
cdk7
|
cyclin-dependent kinase 7 |
| chr19_-_17926712 | 0.49 |
ENSDART00000167592
ENSDART00000162383 |
nkiras1
|
NFKB inhibitor interacting Ras-like 1 |
| chr13_+_33545847 | 0.49 |
ENSDART00000023379
|
mgme1
|
mitochondrial genome maintenance exonuclease 1 |
| chr1_-_40671573 | 0.49 |
ENSDART00000074777
ENSDART00000158114 |
htt
|
huntingtin |
| chr18_+_6498590 | 0.49 |
ENSDART00000162398
|
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr20_-_20711298 | 0.48 |
ENSDART00000063492
|
ppm1ab
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Ab |
| chr22_+_29699600 | 0.48 |
ENSDART00000146280
|
bbip1
|
BBSome interacting protein 1 |
| chr1_+_54270060 | 0.48 |
ENSDART00000152528
|
r3hcc1l
|
R3H domain and coiled-coil containing 1-like |
| chr9_+_38832959 | 0.47 |
ENSDART00000110651
|
slc12a8
|
solute carrier family 12, member 8 |
| chr5_-_66614227 | 0.47 |
ENSDART00000147053
|
rsrc2
|
arginine/serine-rich coiled-coil 2 |
| chr4_-_12979905 | 0.47 |
ENSDART00000150674
|
tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr8_+_48976994 | 0.47 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr25_-_28920272 | 0.46 |
ENSDART00000166889
|
nptna
|
neuroplastin a |
| chr20_-_23354440 | 0.46 |
ENSDART00000103365
|
ociad1
|
OCIA domain containing 1 |
| chr16_+_12922320 | 0.46 |
ENSDART00000008535
|
u2af2a
|
U2 small nuclear RNA auxiliary factor 2a |
| chr13_-_33077267 | 0.45 |
ENSDART00000146138
|
trip11
|
thyroid hormone receptor interactor 11 |
| chr11_+_24110796 | 0.44 |
ENSDART00000089747
|
nfs1
|
NFS1 cysteine desulfurase |
| chr12_-_9430648 | 0.44 |
ENSDART00000152737
ENSDART00000091519 |
pgap3
|
post-GPI attachment to proteins 3 |
| chr23_-_30837818 | 0.43 |
ENSDART00000075918
|
pcmtd2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr14_-_5100304 | 0.43 |
ENSDART00000168074
|
pcgf1
|
polycomb group ring finger 1 |
| chr2_-_42324511 | 0.43 |
ENSDART00000144707
|
slc39a6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr8_-_33145412 | 0.42 |
ENSDART00000133300
|
zbtb34
|
zinc finger and BTB domain containing 34 |
| chr18_-_22745846 | 0.42 |
ENSDART00000023721
|
nudt21
|
nudix hydrolase 21 |
| chr20_-_44562847 | 0.42 |
ENSDART00000085442
|
mut
|
methylmalonyl CoA mutase |
| chr23_-_30837755 | 0.42 |
ENSDART00000075918
|
pcmtd2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr7_+_32358523 | 0.42 |
ENSDART00000138914
|
si:ch73-314g15.3
|
si:ch73-314g15.3 |
| chr16_-_7003373 | 0.42 |
|
|
|
| chr5_+_51186666 | 0.41 |
ENSDART00000165276
|
papd4
|
PAP associated domain containing 4 |
| chr5_+_51186708 | 0.41 |
ENSDART00000165276
|
papd4
|
PAP associated domain containing 4 |
| chr19_-_35155287 | 0.41 |
ENSDART00000175621
|
elp2
|
elongator acetyltransferase complex subunit 2 |
| chr5_-_31889551 | 0.41 |
ENSDART00000168870
|
gpr107
|
G protein-coupled receptor 107 |
| chr19_-_18199573 | 0.40 |
ENSDART00000166313
|
thrb
|
thyroid hormone receptor beta |
| chr20_+_39782876 | 0.40 |
ENSDART00000050729
|
hddc2
|
HD domain containing 2 |
| chr6_-_51666124 | 0.39 |
|
|
|
| chr15_+_19948426 | 0.39 |
ENSDART00000054416
|
dyrk1ab
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, b |
| chr15_+_38397897 | 0.39 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr7_-_50000060 | 0.39 |
ENSDART00000098842
|
hsd17b12b
|
hydroxysteroid (17-beta) dehydrogenase 12b |
| chr6_-_29169308 | 0.39 |
ENSDART00000110288
ENSDART00000008841 |
zbtb11
|
zinc finger and BTB domain containing 11 |
| chr20_-_28939553 | 0.38 |
ENSDART00000044462
|
max
|
myc associated factor X |
| chr20_-_20711525 | 0.38 |
ENSDART00000063492
|
ppm1ab
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Ab |
| chr24_-_32110851 | 0.38 |
ENSDART00000159034
|
rsu1
|
Ras suppressor protein 1 |
| chr22_-_20925422 | 0.38 |
ENSDART00000002029
|
fkbp8
|
FK506 binding protein 8 |
| chr18_-_26815111 | 0.38 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr1_+_11648973 | 0.37 |
ENSDART00000067086
|
nansa
|
N-acetylneuraminic acid synthase a |
| chr24_-_41356967 | 0.37 |
ENSDART00000121592
ENSDART00000019975 ENSDART00000166034 |
acvr2b
|
activin A receptor, type IIB |
| chr7_+_28905846 | 0.36 |
ENSDART00000146655
|
aph1b
|
APH1B gamma secretase subunit |
| chr12_-_31609321 | 0.36 |
ENSDART00000165299
|
srsf2a
|
serine/arginine-rich splicing factor 2a |
| chr24_+_37752868 | 0.36 |
ENSDART00000158181
|
wdr24
|
WD repeat domain 24 |
| chr20_+_39441958 | 0.36 |
ENSDART00000009164
|
esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr6_-_22059932 | 0.35 |
ENSDART00000162148
|
utp6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr16_-_17437374 | 0.34 |
ENSDART00000143056
|
zyx
|
zyxin |
| chr7_-_66470118 | 0.34 |
ENSDART00000021317
|
ctr9
|
CTR9 homolog, Paf1/RNA polymerase II complex component |
| chr23_+_32053034 | 0.34 |
ENSDART00000140351
|
pan2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr21_-_22855247 | 0.34 |
ENSDART00000065563
|
ccdc90b
|
coiled-coil domain containing 90B |
| chr13_-_46424420 | 0.34 |
|
|
|
| chr21_-_30957555 | 0.33 |
ENSDART00000160224
|
pgap2
|
post-GPI attachment to proteins 2 |
| chr24_-_10756995 | 0.33 |
ENSDART00000145593
|
fam49bb
|
family with sequence similarity 49, member Bb |
| chr1_-_40671522 | 0.33 |
ENSDART00000074777
ENSDART00000158114 |
htt
|
huntingtin |
| chr6_-_51666072 | 0.33 |
|
|
|
| chr22_+_520271 | 0.32 |
ENSDART00000067638
|
med20
|
mediator complex subunit 20 |
| chr21_-_14666277 | 0.32 |
ENSDART00000114096
|
arrdc1b
|
arrestin domain containing 1b |
| chr5_-_12586284 | 0.31 |
ENSDART00000051664
|
ypel1
|
yippee-like 1 |
| chr16_+_38387709 | 0.31 |
ENSDART00000135008
|
gabpb2b
|
GA binding protein transcription factor, beta subunit 2b |
| chr12_-_31609517 | 0.31 |
ENSDART00000080173
|
srsf2a
|
serine/arginine-rich splicing factor 2a |
| chr13_+_31415310 | 0.29 |
ENSDART00000164590
|
ppm1aa
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Aa |
| chr12_-_14172962 | 0.28 |
ENSDART00000158399
|
avl9
|
AVL9 homolog (S. cerevisiase) |
| chr7_+_20046811 | 0.28 |
ENSDART00000131019
|
acadvl
|
acyl-CoA dehydrogenase, very long chain |
| chr11_+_25455772 | 0.28 |
ENSDART00000110224
|
mon1bb
|
MON1 secretory trafficking family member Bb |
| chr16_-_44910578 | 0.28 |
ENSDART00000157375
|
arhgap33
|
Rho GTPase activating protein 33 |
| chr25_-_24150270 | 0.28 |
ENSDART00000073527
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr16_+_12922781 | 0.28 |
ENSDART00000008535
|
u2af2a
|
U2 small nuclear RNA auxiliary factor 2a |
| chr20_-_39886349 | 0.27 |
ENSDART00000098253
|
rnf217
|
ring finger protein 217 |
| chr24_-_7797313 | 0.27 |
ENSDART00000112777
|
si:dkey-197c15.6
|
si:dkey-197c15.6 |
| chr8_+_23705243 | 0.26 |
ENSDART00000142395
|
mkrn4
|
makorin, ring finger protein, 4 |
| chr10_+_36495590 | 0.26 |
ENSDART00000124060
|
uspl1
|
ubiquitin specific peptidase like 1 |
| chr24_-_10756925 | 0.26 |
ENSDART00000145593
|
fam49bb
|
family with sequence similarity 49, member Bb |
| chr7_-_66469975 | 0.25 |
ENSDART00000021317
|
ctr9
|
CTR9 homolog, Paf1/RNA polymerase II complex component |
| chr15_+_19948680 | 0.25 |
ENSDART00000054416
|
dyrk1ab
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, b |
| chr1_-_51086075 | 0.25 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr19_-_43468824 | 0.25 |
|
|
|
| chr22_-_18466730 | 0.25 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr17_+_24666346 | 0.24 |
ENSDART00000146309
|
znf593
|
zinc finger protein 593 |
| chr21_+_37760978 | 0.24 |
ENSDART00000145828
|
pgk1
|
phosphoglycerate kinase 1 |
| chr19_+_43468926 | 0.23 |
|
|
|
| chr12_-_25006119 | 0.23 |
ENSDART00000158036
|
cript
|
cysteine-rich PDZ-binding protein |
| chr23_+_39454172 | 0.23 |
ENSDART00000149819
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr24_-_32110808 | 0.22 |
ENSDART00000159034
|
rsu1
|
Ras suppressor protein 1 |
| chr24_-_10756953 | 0.22 |
ENSDART00000145593
|
fam49bb
|
family with sequence similarity 49, member Bb |
| chr15_+_17407258 | 0.22 |
ENSDART00000111753
|
vmp1
|
vacuole membrane protein 1 |
| chr2_+_52225552 | 0.22 |
ENSDART00000170353
|
ENSDARG00000103108
|
ENSDARG00000103108 |
| chr12_+_31307634 | 0.22 |
ENSDART00000153179
|
zdhhc6
|
zinc finger, DHHC-type containing 6 |
| chr9_-_28444798 | 0.21 |
|
|
|
| chr18_-_18595722 | 0.21 |
ENSDART00000159274
|
sf3b3
|
splicing factor 3b, subunit 3 |
| chr5_-_68712571 | 0.20 |
ENSDART00000109487
|
CABZ01032476.1
|
ENSDARG00000074645 |
| chr7_+_28906011 | 0.20 |
ENSDART00000008096
|
aph1b
|
APH1B gamma secretase subunit |
| chr14_-_23387483 | 0.20 |
ENSDART00000024604
|
mars2
|
methionyl-tRNA synthetase 2, mitochondrial |
| chr7_+_72030256 | 0.19 |
ENSDART00000172021
|
tollip
|
toll interacting protein |
| chr20_-_9793709 | 0.19 |
ENSDART00000053838
|
cgrrf1
|
cell growth regulator with ring finger domain 1 |
| chr8_+_48976926 | 0.19 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr8_+_24721104 | 0.18 |
ENSDART00000126897
|
lamtor5
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
| chr15_-_14706119 | 0.18 |
ENSDART00000164166
|
si:dkey-260j18.2
|
si:dkey-260j18.2 |
| chr1_+_54270383 | 0.17 |
ENSDART00000152528
|
r3hcc1l
|
R3H domain and coiled-coil containing 1-like |
| chr7_-_19851755 | 0.17 |
ENSDART00000052902
|
acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr3_-_36277628 | 0.17 |
ENSDART00000173545
|
cog1
|
component of oligomeric golgi complex 1 |
| chr19_+_35155362 | 0.17 |
ENSDART00000146777
ENSDART00000103276 |
fam206a
|
family with sequence similarity 206, member A |
| chr7_-_19851362 | 0.17 |
ENSDART00000052902
|
acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr7_-_33791033 | 0.16 |
ENSDART00000052404
|
map2k5
|
mitogen-activated protein kinase kinase 5 |
| chr12_-_14173331 | 0.16 |
ENSDART00000158399
|
avl9
|
AVL9 homolog (S. cerevisiase) |
| chr21_+_37985488 | 0.16 |
ENSDART00000085728
|
klf8
|
Kruppel-like factor 8 |
| chr1_-_28146457 | 0.15 |
ENSDART00000110270
ENSDART00000170797 |
pwp2h
|
PWP2 periodic tryptophan protein homolog (yeast) |
| chr2_+_2930605 | 0.15 |
ENSDART00000124882
|
thoc1
|
THO complex 1 |
| chr20_-_44562723 | 0.15 |
ENSDART00000085442
|
mut
|
methylmalonyl CoA mutase |
| chr2_-_42324343 | 0.15 |
ENSDART00000098357
|
slc39a6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr7_-_13661739 | 0.15 |
ENSDART00000062257
|
slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr19_-_27422599 | 0.15 |
ENSDART00000089540
|
sacm1la
|
SAC1 like phosphatidylinositide phosphatase a |
| chr12_-_14172723 | 0.15 |
ENSDART00000077903
ENSDART00000163758 |
avl9
|
AVL9 homolog (S. cerevisiase) |
| chr25_+_3221887 | 0.15 |
ENSDART00000104888
|
slc35b4
|
solute carrier family 35, member B4 |
| chr5_+_41463969 | 0.15 |
ENSDART00000035235
|
UBB
|
si:ch211-202a12.4 |
| chr12_-_9430818 | 0.14 |
ENSDART00000152737
ENSDART00000091519 |
pgap3
|
post-GPI attachment to proteins 3 |
| chr21_+_37985536 | 0.14 |
ENSDART00000085728
|
klf8
|
Kruppel-like factor 8 |
| chr15_-_29181452 | 0.14 |
ENSDART00000145748
ENSDART00000109482 ENSDART00000179123 |
ENSDARG00000074967
|
ENSDARG00000074967 |
| chr5_-_68712851 | 0.13 |
ENSDART00000109487
|
CABZ01032476.1
|
ENSDARG00000074645 |
| chr16_+_33984153 | 0.13 |
ENSDART00000166254
|
gpn2
|
GPN-loop GTPase 2 |
| chr13_+_13875110 | 0.13 |
ENSDART00000131875
|
atrn
|
attractin |
| chr16_+_12922149 | 0.13 |
ENSDART00000124875
|
u2af2a
|
U2 small nuclear RNA auxiliary factor 2a |
| chr21_-_14666223 | 0.13 |
ENSDART00000114096
|
arrdc1b
|
arrestin domain containing 1b |
| chr6_+_41811132 | 0.12 |
ENSDART00000038163
|
rad18
|
RAD18 E3 ubiquitin protein ligase |
| chr17_+_43478181 | 0.12 |
ENSDART00000055487
|
chmp3
|
charged multivesicular body protein 3 |
| chr23_-_27621359 | 0.12 |
ENSDART00000144419
|
larp4aa
|
La ribonucleoprotein domain family, member 4Aa |
| chr22_+_520319 | 0.12 |
ENSDART00000067638
|
med20
|
mediator complex subunit 20 |
| chr24_-_37752716 | 0.12 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr23_-_30837699 | 0.12 |
ENSDART00000075918
|
pcmtd2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr21_-_30957754 | 0.11 |
ENSDART00000065503
|
pgap2
|
post-GPI attachment to proteins 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0031439 | pro-B cell differentiation(GO:0002328) regulation of mRNA cleavage(GO:0031437) positive regulation of mRNA cleavage(GO:0031439) regulation of pro-B cell differentiation(GO:2000973) positive regulation of pro-B cell differentiation(GO:2000975) |
| 0.6 | 1.7 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.3 | 1.9 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.3 | 0.8 | GO:0090017 | neural plate formation(GO:0021990) brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) anterior neural plate formation(GO:0090017) |
| 0.2 | 1.2 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.2 | 2.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.2 | 0.5 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.2 | 0.5 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.2 | 0.5 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.2 | 0.6 | GO:0042779 | tRNA 3'-trailer cleavage(GO:0042779) |
| 0.1 | 0.7 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.1 | 0.6 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.1 | 0.4 | GO:0046552 | thyroid hormone mediated signaling pathway(GO:0002154) retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.1 | 0.4 | GO:0042128 | nitrate metabolic process(GO:0042126) nitrate assimilation(GO:0042128) |
| 0.1 | 0.4 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.7 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 1.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 1.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.6 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.2 | GO:0098751 | bone cell development(GO:0098751) |
| 0.1 | 1.3 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 0.3 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 0.5 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.4 | GO:0008210 | estrogen biosynthetic process(GO:0006703) estrogen metabolic process(GO:0008210) |
| 0.1 | 0.9 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.5 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) regulation of lamellipodium organization(GO:1902743) |
| 0.1 | 0.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.9 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 0.3 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.1 | 0.6 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 0.5 | GO:0048512 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 0.2 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 0.8 | GO:0050870 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.7 | GO:0046660 | female sex differentiation(GO:0046660) |
| 0.0 | 0.1 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.0 | 1.8 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.5 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.4 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.9 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.6 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.5 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.5 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.8 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.4 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.4 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.8 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.5 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 0.2 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.1 | GO:0071850 | positive regulation of cytokinesis(GO:0032467) mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.5 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.4 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.3 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.3 | 1.9 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 1.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 0.7 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 0.7 | GO:0098536 | deuterosome(GO:0098536) |
| 0.2 | 1.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.5 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 1.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.6 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 0.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.7 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 0.3 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.6 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.8 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.3 | 2.8 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 0.6 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.2 | 0.6 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.2 | 1.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.4 | GO:0046857 | nitrite reductase [NAD(P)H] activity(GO:0008942) oxidoreductase activity, acting on other nitrogenous compounds as donors, with NAD or NADP as acceptor(GO:0046857) |
| 0.1 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.4 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 0.5 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.1 | 2.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.7 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 2.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 0.7 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.6 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.1 | 1.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 2.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.2 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.1 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.4 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.4 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.5 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.2 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.8 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 1.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.5 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.4 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.6 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 0.2 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.1 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.0 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.3 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.4 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.2 | 1.8 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.1 | 2.8 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |