Project
DANIO-CODE
Navigation
Downloads
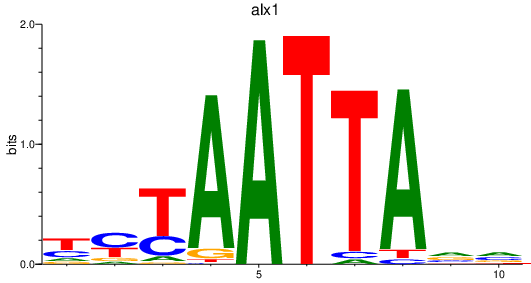
Results for alx1
Z-value: 0.95
Transcription factors associated with alx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
alx1
|
ENSDARG00000062824 | ALX homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| alx1 | dr10_dc_chr18_+_16257606_16257627 | -0.80 | 2.1e-04 | Click! |
Activity profile of alx1 motif
Sorted Z-values of alx1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of alx1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_15536640 | 5.19 |
ENSDART00000098970
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr8_+_45326435 | 4.53 |
ENSDART00000134161
|
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr10_-_25246786 | 3.34 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr5_+_37303599 | 3.34 |
ENSDART00000097754
ENSDART00000162470 |
tmprss4b
|
transmembrane protease, serine 4b |
| chr5_+_57254393 | 3.30 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr21_+_25740782 | 3.30 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr10_-_21404605 | 3.22 |
ENSDART00000125167
|
avd
|
avidin |
| chr12_-_14104939 | 3.02 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr7_-_51497945 | 3.00 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr11_-_1524107 | 2.90 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr19_-_20819477 | 2.80 |
ENSDART00000151356
|
dazl
|
deleted in azoospermia-like |
| chr16_+_39209567 | 2.70 |
ENSDART00000121756
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr14_+_34150130 | 2.65 |
ENSDART00000132193
ENSDART00000141058 |
wnt8a
BX927327.1
|
wingless-type MMTV integration site family, member 8a ENSDARG00000105311 |
| chr24_-_14446593 | 2.61 |
|
|
|
| chr24_+_12689711 | 2.59 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr3_+_32285237 | 2.57 |
ENSDART00000157324
|
prrg2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr10_-_34971985 | 2.54 |
ENSDART00000141201
|
ccna1
|
cyclin A1 |
| chr16_-_29452509 | 2.50 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr22_-_17627900 | 2.49 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr18_-_43890836 | 2.31 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr19_-_20819101 | 2.29 |
ENSDART00000137590
|
dazl
|
deleted in azoospermia-like |
| chr17_+_16038358 | 2.28 |
ENSDART00000155336
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr10_+_6925373 | 2.12 |
ENSDART00000128866
|
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr11_-_34909095 | 2.12 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr24_+_8702288 | 2.05 |
ENSDART00000114810
|
sycp2l
|
synaptonemal complex protein 2-like |
| chr16_-_46613217 | 1.89 |
ENSDART00000127212
|
si:dkey-152b24.6
|
si:dkey-152b24.6 |
| chr24_+_19270877 | 1.87 |
|
|
|
| chr1_-_54570813 | 1.83 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr25_+_7261050 | 1.83 |
ENSDART00000163017
|
prc1a
|
protein regulator of cytokinesis 1a |
| chr11_-_6442588 | 1.78 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr20_-_23527234 | 1.73 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr17_+_16038103 | 1.73 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| KN149710v1_+_38638 | 1.72 |
|
|
|
| chr19_-_5186692 | 1.72 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr11_+_34909167 | 1.69 |
ENSDART00000110839
|
mon1a
|
MON1 secretory trafficking family member A |
| chr16_-_39209426 | 1.69 |
|
|
|
| chr14_-_8634381 | 1.69 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
| chr5_+_58936399 | 1.68 |
|
|
|
| chr11_+_34909244 | 1.68 |
ENSDART00000110839
|
mon1a
|
MON1 secretory trafficking family member A |
| chr16_+_42567707 | 1.60 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr11_+_23799984 | 1.58 |
|
|
|
| chr1_-_18118467 | 1.57 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr22_-_17627831 | 1.45 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr16_-_29451902 | 1.45 |
|
|
|
| chr18_+_20571460 | 1.44 |
ENSDART00000151990
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr10_-_25448712 | 1.44 |
ENSDART00000140023
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr10_+_19625897 | 1.43 |
|
|
|
| chr24_+_12689887 | 1.43 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr8_+_41003546 | 1.42 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr5_-_8712114 | 1.42 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr15_-_23484927 | 1.42 |
ENSDART00000148840
|
kmt2a
|
lysine (K)-specific methyltransferase 2A |
| chr12_-_13511174 | 1.41 |
ENSDART00000133895
|
ghdc
|
GH3 domain containing |
| chr10_-_34971926 | 1.40 |
ENSDART00000141201
|
ccna1
|
cyclin A1 |
| chr11_-_40154790 | 1.39 |
ENSDART00000086296
|
trim62
|
tripartite motif containing 62 |
| chr7_-_24604255 | 1.38 |
ENSDART00000173920
|
adad2
|
adenosine deaminase domain containing 2 |
| chr7_+_28341426 | 1.36 |
ENSDART00000019991
|
slc7a6os
|
solute carrier family 7, member 6 opposite strand |
| chr24_-_30929990 | 1.35 |
|
|
|
| chr1_+_50547385 | 1.32 |
ENSDART00000132141
|
btbd3a
|
BTB (POZ) domain containing 3a |
| chr22_+_4035577 | 1.32 |
ENSDART00000170620
|
ctxn1
|
cortexin 1 |
| chr6_+_29702868 | 1.30 |
ENSDART00000114172
|
pde6d
|
phosphodiesterase 6D, cGMP-specific, rod, delta |
| chr2_-_26941084 | 1.30 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr1_-_7160294 | 1.29 |
ENSDART00000174972
|
BX908756.2
|
ENSDARG00000108483 |
| chr2_+_6341404 | 1.27 |
ENSDART00000076700
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr8_+_49128711 | 1.27 |
ENSDART00000079631
|
rad21l1
|
RAD21 cohesin complex component like 1 |
| chr21_-_32027717 | 1.26 |
ENSDART00000131651
|
ENSDARG00000073961
|
ENSDARG00000073961 |
| chr20_+_16740320 | 1.25 |
ENSDART00000152359
|
tmem30ab
|
transmembrane protein 30Ab |
| chr16_+_33209956 | 1.24 |
ENSDART00000101943
|
rragca
|
Ras-related GTP binding Ca |
| chr7_+_24855467 | 1.23 |
ENSDART00000077217
|
zgc:101765
|
zgc:101765 |
| chr8_+_25126238 | 1.23 |
ENSDART00000136505
|
gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr15_+_35089305 | 1.23 |
ENSDART00000156515
|
zgc:55621
|
zgc:55621 |
| chr13_+_24549364 | 1.22 |
ENSDART00000139854
|
zgc:66426
|
zgc:66426 |
| chr20_+_32620786 | 1.22 |
ENSDART00000147319
|
scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr20_+_19304349 | 1.20 |
|
|
|
| chr5_-_8712068 | 1.19 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr9_+_29737843 | 1.19 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr7_+_22042296 | 1.18 |
ENSDART00000123457
|
TMEM102
|
transmembrane protein 102 |
| chr16_+_19831573 | 1.18 |
ENSDART00000135359
|
macc1
|
metastasis associated in colon cancer 1 |
| chr8_+_28362546 | 1.18 |
ENSDART00000062682
|
adipor1b
|
adiponectin receptor 1b |
| chr3_+_28729443 | 1.18 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr24_-_10935904 | 1.17 |
ENSDART00000003195
|
chmp4c
|
charged multivesicular body protein 4C |
| chr6_+_40925259 | 1.17 |
ENSDART00000002728
ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr14_+_25950244 | 1.14 |
ENSDART00000113804
ENSDART00000159054 |
CCDC69
|
coiled-coil domain containing 69 |
| chr18_-_50439653 | 1.13 |
ENSDART00000151038
|
CU862078.1
|
ENSDARG00000096262 |
| chr2_+_23383492 | 1.13 |
ENSDART00000170669
|
csnk1g2a
|
casein kinase 1, gamma 2a |
| chr1_+_35253862 | 1.11 |
ENSDART00000139636
|
zgc:152968
|
zgc:152968 |
| chr19_+_38262119 | 1.11 |
ENSDART00000042276
|
nxph1
|
neurexophilin 1 |
| chr25_+_29219342 | 1.10 |
ENSDART00000108692
ENSDART00000077445 |
pim3
|
Pim-3 proto-oncogene, serine/threonine kinase |
| chr13_+_22973764 | 1.09 |
ENSDART00000110266
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr18_+_20571400 | 1.09 |
ENSDART00000151990
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr10_+_17277353 | 1.09 |
ENSDART00000038780
|
sppl3
|
signal peptide peptidase 3 |
| chr21_+_31216418 | 1.08 |
ENSDART00000178521
|
asl
|
argininosuccinate lyase |
| chr17_+_26704439 | 1.07 |
ENSDART00000114927
|
nrde2
|
NRDE-2, necessary for RNA interference, domain containing |
| chr16_+_29574449 | 1.07 |
ENSDART00000148450
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr22_-_551324 | 1.07 |
|
|
|
| chr5_+_6391432 | 1.05 |
ENSDART00000170564
ENSDART00000086666 |
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr14_-_33141111 | 1.04 |
ENSDART00000147059
|
lamp2
|
lysosomal-associated membrane protein 2 |
| chr13_-_32595667 | 1.03 |
ENSDART00000012232
|
pdss2
|
prenyl (decaprenyl) diphosphate synthase, subunit 2 |
| chr15_-_16241412 | 1.02 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| KN150065v1_-_622 | 1.02 |
|
|
|
| chr14_-_26199859 | 1.01 |
ENSDART00000054175
ENSDART00000145625 |
smad5
|
SMAD family member 5 |
| chr12_-_30244575 | 1.01 |
ENSDART00000152981
|
tdrd1
|
tudor domain containing 1 |
| chr14_+_31265616 | 1.01 |
|
|
|
| chr13_-_32768928 | 1.01 |
ENSDART00000163757
|
rock2a
|
rho-associated, coiled-coil containing protein kinase 2a |
| chr21_-_36710989 | 1.00 |
ENSDART00000086060
|
mrpl22
|
mitochondrial ribosomal protein L22 |
| chr13_-_32595706 | 1.00 |
ENSDART00000012232
|
pdss2
|
prenyl (decaprenyl) diphosphate synthase, subunit 2 |
| chr2_+_30013468 | 1.00 |
ENSDART00000139566
|
rbm33b
|
RNA binding motif protein 33b |
| chr13_-_25590425 | 1.00 |
ENSDART00000142404
|
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr20_+_54404987 | 0.99 |
ENSDART00000099338
|
actr10
|
ARP10 actin related protein 10 homolog |
| chr14_-_21816253 | 0.99 |
ENSDART00000113752
|
si:dkey-6i22.5
|
si:dkey-6i22.5 |
| chr22_+_1153361 | 0.99 |
ENSDART00000159761
|
irf6
|
interferon regulatory factor 6 |
| chr2_-_27892824 | 0.98 |
|
|
|
| chr22_+_17803309 | 0.98 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr11_+_38858311 | 0.98 |
ENSDART00000155746
|
cdc42
|
cell division cycle 42 |
| chr20_+_32620937 | 0.98 |
ENSDART00000147319
|
scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr23_-_35691369 | 0.98 |
ENSDART00000142369
|
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr21_+_34053739 | 0.97 |
ENSDART00000147519
|
mtmr1b
|
myotubularin related protein 1b |
| chr2_-_26941232 | 0.97 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr8_+_11387135 | 0.96 |
|
|
|
| chr5_-_61134500 | 0.95 |
ENSDART00000079855
|
im:7138535
|
im:7138535 |
| chr6_-_41031013 | 0.94 |
ENSDART00000134293
|
trub2
|
TruB pseudouridine (psi) synthase family member 2 |
| chr8_+_19945820 | 0.94 |
ENSDART00000134124
|
znf692
|
zinc finger protein 692 |
| chr12_-_30244042 | 0.94 |
ENSDART00000152878
|
tdrd1
|
tudor domain containing 1 |
| chr23_+_28464194 | 0.94 |
ENSDART00000133736
|
CT025585.1
|
ENSDARG00000093306 |
| chr8_-_22720007 | 0.93 |
|
|
|
| chr20_-_37910887 | 0.92 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr8_-_1833095 | 0.90 |
ENSDART00000114476
ENSDART00000091235 ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr18_+_19467527 | 0.90 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr15_+_36075206 | 0.90 |
ENSDART00000019976
|
rhbdd1
|
rhomboid domain containing 1 |
| chr21_-_25765319 | 0.90 |
ENSDART00000101219
|
mettl27
|
methyltransferase like 27 |
| chr16_-_32865452 | 0.89 |
|
|
|
| chr16_+_47283253 | 0.89 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr13_-_36015558 | 0.88 |
ENSDART00000133072
|
med6
|
mediator complex subunit 6 |
| chr16_-_25452949 | 0.87 |
ENSDART00000167574
|
rbfa
|
ribosome binding factor A |
| chr7_+_12675166 | 0.87 |
ENSDART00000027329
|
fth1a
|
ferritin, heavy polypeptide 1a |
| chr21_-_14154489 | 0.87 |
ENSDART00000114715
|
man1b1a
|
mannosidase, alpha, class 1B, member 1a |
| chr23_+_32102030 | 0.85 |
ENSDART00000145501
|
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr1_+_8010026 | 0.85 |
ENSDART00000152142
|
zgc:77849
|
zgc:77849 |
| chr3_-_40695299 | 0.84 |
ENSDART00000134026
|
wipi2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr23_-_43916621 | 0.83 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr17_+_25425914 | 0.82 |
ENSDART00000030691
|
clic4
|
chloride intracellular channel 4 |
| chr24_+_16924542 | 0.82 |
ENSDART00000014787
|
pip4k2aa
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a |
| chr19_-_20819057 | 0.82 |
ENSDART00000136826
|
dazl
|
deleted in azoospermia-like |
| chr9_+_35205519 | 0.81 |
ENSDART00000100700
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr8_-_44247277 | 0.81 |
|
|
|
| chr8_+_50964745 | 0.81 |
ENSDART00000013870
|
ENSDARG00000007359
|
ENSDARG00000007359 |
| chr1_+_50547341 | 0.81 |
ENSDART00000132141
|
btbd3a
|
BTB (POZ) domain containing 3a |
| chr25_-_28630138 | 0.80 |
|
|
|
| chr16_+_35448634 | 0.80 |
ENSDART00000171608
|
rab42b
|
RAB42, member RAS oncogene family |
| chr20_+_27341244 | 0.80 |
ENSDART00000123950
|
ENSDARG00000077377
|
ENSDARG00000077377 |
| chr24_-_2454189 | 0.80 |
ENSDART00000093331
|
rreb1a
|
ras responsive element binding protein 1a |
| chr1_+_18118735 | 0.79 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr6_+_48188549 | 0.78 |
|
|
|
| chr24_-_30930178 | 0.78 |
|
|
|
| chr16_+_33209755 | 0.78 |
ENSDART00000101943
|
rragca
|
Ras-related GTP binding Ca |
| chr2_-_57826189 | 0.78 |
ENSDART00000131420
|
si:dkeyp-68b7.5
|
si:dkeyp-68b7.5 |
| chr10_-_32550351 | 0.77 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr15_+_21327206 | 0.77 |
ENSDART00000101000
|
gkup
|
glucuronokinase with putative uridyl pyrophosphorylase |
| chr6_+_50382153 | 0.77 |
ENSDART00000055504
|
cyc1
|
cytochrome c-1 |
| chr14_-_26093969 | 0.77 |
ENSDART00000037999
|
b4galt7
|
xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 (galactosyltransferase I) |
| chr10_+_43953171 | 0.77 |
|
|
|
| chr3_-_40695407 | 0.77 |
ENSDART00000134026
|
wipi2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr18_+_5066229 | 0.76 |
ENSDART00000157671
|
CABZ01080599.1
|
ENSDARG00000100626 |
| chr24_-_21788942 | 0.76 |
ENSDART00000113092
|
abhd10b
|
abhydrolase domain containing 10b |
| chr18_+_8362265 | 0.74 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr13_+_31271568 | 0.73 |
ENSDART00000019202
|
tdrd9
|
tudor domain containing 9 |
| chr19_-_18664670 | 0.73 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr16_+_39209538 | 0.73 |
ENSDART00000121756
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr24_-_30929960 | 0.73 |
|
|
|
| chr21_+_34053590 | 0.73 |
ENSDART00000147519
ENSDART00000158115 ENSDART00000029599 ENSDART00000145123 |
mtmr1b
|
myotubularin related protein 1b |
| chr20_+_32620873 | 0.73 |
ENSDART00000147319
|
scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr5_+_37303707 | 0.72 |
ENSDART00000097754
ENSDART00000162470 |
tmprss4b
|
transmembrane protease, serine 4b |
| chr3_+_37507517 | 0.72 |
ENSDART00000075039
|
gosr2
|
golgi SNAP receptor complex member 2 |
| chr18_+_20571785 | 0.72 |
ENSDART00000040074
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr2_-_38380883 | 0.72 |
ENSDART00000088026
|
prmt5
|
protein arginine methyltransferase 5 |
| chr20_-_16271738 | 0.72 |
ENSDART00000012476
|
coa7
|
cytochrome c oxidase assembly factor 7 |
| chr6_-_12665211 | 0.72 |
ENSDART00000150887
|
ical1
|
islet cell autoantigen 1-like |
| chr18_+_27769446 | 0.72 |
|
|
|
| chr2_-_26940965 | 0.71 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr16_+_47283374 | 0.71 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr16_-_12582192 | 0.71 |
|
|
|
| chr5_+_25133592 | 0.71 |
ENSDART00000098467
|
abhd17b
|
abhydrolase domain containing 17B |
| chr2_-_22980899 | 0.71 |
ENSDART00000056963
|
stk25b
|
serine/threonine kinase 25b |
| chr8_+_45326523 | 0.70 |
ENSDART00000145011
|
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr4_-_66538843 | 0.70 |
ENSDART00000165173
|
BX548011.1
|
ENSDARG00000103357 |
| chr21_-_13026036 | 0.70 |
ENSDART00000135623
|
fam219aa
|
family with sequence similarity 219, member Aa |
| chr19_+_40482359 | 0.69 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr19_+_42657913 | 0.69 |
ENSDART00000131574
ENSDART00000135436 |
jtb
|
jumping translocation breakpoint |
| chr17_+_16082165 | 0.69 |
ENSDART00000132203
|
znf395a
|
zinc finger protein 395a |
| chr14_+_23420053 | 0.69 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr23_+_28464298 | 0.69 |
ENSDART00000133736
|
CT025585.1
|
ENSDARG00000093306 |
| chr15_-_43402935 | 0.68 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr16_+_42567668 | 0.68 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr23_+_17583666 | 0.68 |
ENSDART00000054738
|
gid8b
|
GID complex subunit 8 homolog b (S. cerevisiae) |
| chr24_-_14447519 | 0.68 |
|
|
|
| chr5_-_19987344 | 0.68 |
ENSDART00000144232
|
ficd
|
FIC domain containing |
| chr23_+_28464143 | 0.68 |
ENSDART00000133736
|
CT025585.1
|
ENSDARG00000093306 |
| chr18_+_36160190 | 0.68 |
ENSDART00000146068
|
BX927288.1
|
ENSDARG00000092309 |
| chr3_-_39554099 | 0.67 |
ENSDART00000145303
|
b9d1
|
B9 protein domain 1 |
| chr13_-_35346618 | 0.66 |
ENSDART00000057052
|
slx4ip
|
SLX4 interacting protein |
| chr21_-_36768113 | 0.66 |
ENSDART00000018350
|
cnot8
|
CCR4-NOT transcription complex, subunit 8 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.9 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 1.2 | 4.6 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.8 | 3.4 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.8 | 3.0 | GO:0042698 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.7 | 2.0 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.6 | 3.2 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.6 | 1.7 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.5 | 1.5 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.4 | 4.4 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.4 | 2.1 | GO:0048608 | gonad development(GO:0008406) reproductive structure development(GO:0048608) reproductive system development(GO:0061458) |
| 0.3 | 3.1 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.3 | 1.0 | GO:0060775 | protein catabolic process in the vacuole(GO:0007039) mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) |
| 0.3 | 1.7 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.3 | 3.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 0.9 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 0.8 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.2 | 0.6 | GO:1903895 | regulation of endoplasmic reticulum unfolded protein response(GO:1900101) negative regulation of endoplasmic reticulum unfolded protein response(GO:1900102) regulation of IRE1-mediated unfolded protein response(GO:1903894) negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.2 | 1.0 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.2 | 0.6 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.2 | 0.7 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.2 | 1.8 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.2 | 0.9 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.2 | 1.5 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.2 | 1.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 6.1 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.2 | 1.3 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.2 | 0.9 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.2 | 0.9 | GO:1902425 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) regulation of mitotic attachment of spindle microtubules to kinetochore(GO:1902423) positive regulation of attachment of mitotic spindle microtubules to kinetochore(GO:1902425) RNA localization to chromatin(GO:1990280) |
| 0.1 | 0.3 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 1.6 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 2.2 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.6 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 1.8 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.1 | 3.3 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 1.7 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.1 | 1.1 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.1 | 1.4 | GO:0048339 | paraxial mesoderm development(GO:0048339) |
| 0.1 | 0.5 | GO:0030238 | male sex determination(GO:0030238) androst-4-ene-3,17-dione biosynthetic process(GO:1903449) |
| 0.1 | 0.3 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) establishment of mitochondrion localization(GO:0051654) |
| 0.1 | 1.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.6 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 1.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.6 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 2.0 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 1.2 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 0.7 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 0.9 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.1 | 0.6 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.6 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) regulation of postsynaptic membrane potential(GO:0060078) |
| 0.1 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 4.3 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 0.1 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 0.7 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.2 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.1 | 0.2 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 0.2 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.1 | 0.6 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 3.5 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.1 | 2.0 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.1 | 0.3 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.4 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.1 | 0.3 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.5 | GO:0046386 | deoxyribonucleotide catabolic process(GO:0009264) deoxyribose phosphate catabolic process(GO:0046386) |
| 0.1 | 0.6 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.1 | 2.1 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.1 | 1.8 | GO:0048599 | oocyte development(GO:0048599) |
| 0.1 | 1.4 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 0.4 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.3 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.7 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.8 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.9 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 4.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 3.8 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.1 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.3 | GO:2001287 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.5 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 1.1 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.2 | GO:0007063 | regulation of sister chromatid cohesion(GO:0007063) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.9 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.3 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 1.2 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 1.5 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.4 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.2 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.4 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 1.7 | GO:0007346 | regulation of mitotic cell cycle(GO:0007346) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 1.4 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.1 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 1.3 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.7 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 1.9 | GO:0015698 | inorganic anion transport(GO:0015698) |
| 0.0 | 1.0 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 1.4 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.4 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) negative regulation of protein modification by small protein conjugation or removal(GO:1903321) |
| 0.0 | 0.3 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 1.2 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 0.1 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.4 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.8 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 0.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.3 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.5 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.3 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.2 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.4 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 0.5 | GO:0018198 | peptidyl-cysteine modification(GO:0018198) |
| 0.0 | 0.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.3 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 0.8 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.3 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.2 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.3 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.1 | GO:1902766 | wound healing, spreading of cells(GO:0044319) epiboly involved in wound healing(GO:0090505) skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.5 | GO:0003341 | cilium movement(GO:0003341) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.8 | 3.4 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.4 | 1.3 | GO:0034990 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.3 | 1.0 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.3 | 2.4 | GO:0071546 | pi-body(GO:0071546) |
| 0.3 | 1.0 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.3 | 4.7 | GO:0043186 | P granule(GO:0043186) |
| 0.3 | 2.0 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 1.2 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.2 | 1.1 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 6.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 2.7 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.2 | 1.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 1.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 3.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.9 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 1.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.9 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.6 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.6 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 1.6 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 1.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.7 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 1.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.4 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.4 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.5 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 1.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.3 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 1.4 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.8 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 2.7 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 1.3 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.8 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 1.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 2.7 | GO:0005815 | microtubule organizing center(GO:0005815) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.6 | 1.7 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.5 | 5.9 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.5 | 3.2 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.5 | 2.0 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.5 | 1.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.5 | 5.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.3 | 1.3 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 0.3 | 2.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.3 | 1.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 3.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 0.9 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.2 | 0.8 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.2 | 3.3 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.2 | 1.8 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 0.6 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.2 | 1.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 2.0 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.2 | 1.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 0.6 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.2 | 0.9 | GO:0034046 | TFIIH-class transcription factor binding(GO:0001097) poly(C) RNA binding(GO:0017130) poly(G) binding(GO:0034046) |
| 0.2 | 0.9 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 1.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.7 | GO:0046922 | peptide-O-fucosyltransferase activity(GO:0046922) |
| 0.1 | 4.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.5 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.1 | 0.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.0 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 0.9 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.9 | GO:0016724 | ferroxidase activity(GO:0004322) ferric iron binding(GO:0008199) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.1 | 0.6 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 2.1 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 0.3 | GO:1990757 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.1 | 1.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.4 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.1 | 0.5 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 3.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 1.6 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 4.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 0.3 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 0.4 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.1 | 4.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.9 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 1.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 0.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 2.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.4 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 0.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.3 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.6 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 1.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.9 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 6.9 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.5 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.5 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.8 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 1.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.5 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.5 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.7 | GO:0015103 | inorganic anion transmembrane transporter activity(GO:0015103) |
| 0.0 | 2.7 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.8 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 2.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.3 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 1.2 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.5 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.3 | 1.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.3 | 2.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 3.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 0.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 6.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.6 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.2 | 3.9 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 0.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 0.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.6 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 1.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.9 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.0 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.7 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 0.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |