Project
DANIO-CODE
Navigation
Downloads
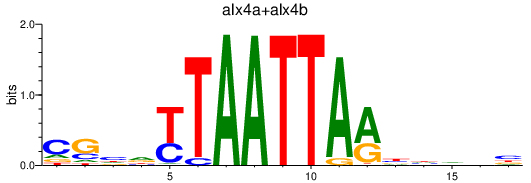
Results for alx4a+alx4b
Z-value: 0.73
Transcription factors associated with alx4a+alx4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
alx4b
|
ENSDARG00000074442 | ALX homeobox 4b |
|
alx4a
|
ENSDARG00000088332 | ALX homeobox 4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| alx4a | dr10_dc_chr7_-_26653629_26653637 | 0.21 | 4.4e-01 | Click! |
Activity profile of alx4a+alx4b motif
Sorted Z-values of alx4a+alx4b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of alx4a+alx4b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_6289363 | 0.62 |
ENSDART00000138600
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr5_+_39885307 | 0.52 |
ENSDART00000051055
|
ndufs4
|
NADH dehydrogenase (ubiquinone) Fe-S protein 4, (NADH-coenzyme Q reductase) |
| chr5_-_21015548 | 0.51 |
ENSDART00000040184
|
tenm1
|
teneurin transmembrane protein 1 |
| chr3_+_50511676 | 0.51 |
ENSDART00000102202
|
ppap2d
|
phosphatidic acid phosphatase type 2D |
| chr8_+_44619220 | 0.50 |
ENSDART00000063392
|
lsm1
|
LSM1, U6 small nuclear RNA associated |
| chr7_-_29906486 | 0.49 |
ENSDART00000046689
|
tmed3
|
transmembrane p24 trafficking protein 3 |
| chr5_-_3673698 | 0.47 |
|
|
|
| chr4_-_1952201 | 0.46 |
ENSDART00000135749
|
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr2_-_17721575 | 0.45 |
ENSDART00000141188
ENSDART00000100201 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr17_+_43041951 | 0.44 |
ENSDART00000154863
|
isca2
|
iron-sulfur cluster assembly 2 |
| chr2_+_25659945 | 0.44 |
ENSDART00000161386
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr16_+_23516127 | 0.43 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr18_+_8362131 | 0.43 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr8_+_29733109 | 0.43 |
ENSDART00000020621
|
mapk4
|
mitogen-activated protein kinase 4 |
| chr12_+_47134830 | 0.42 |
ENSDART00000169406
ENSDART00000158568 ENSDART00000160616 |
mtr
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr19_+_1743359 | 0.41 |
ENSDART00000166744
|
dennd3a
|
DENN/MADD domain containing 3a |
| chr4_-_1952230 | 0.41 |
ENSDART00000135749
|
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr25_+_4454862 | 0.40 |
ENSDART00000110598
|
zmp:0000001167
|
zmp:0000001167 |
| chr6_+_11014565 | 0.39 |
ENSDART00000132677
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr13_-_36672606 | 0.39 |
ENSDART00000179242
|
sav1
|
salvador family WW domain containing protein 1 |
| chr18_+_39506453 | 0.38 |
ENSDART00000126978
|
acadl
|
acyl-CoA dehydrogenase, long chain |
| chr23_-_24556810 | 0.38 |
ENSDART00000109248
|
spen
|
spen family transcriptional repressor |
| chr8_+_31426328 | 0.38 |
ENSDART00000135101
|
sepp1a
|
selenoprotein P, plasma, 1a |
| chr3_-_32686790 | 0.37 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr14_+_5078937 | 0.35 |
ENSDART00000031508
|
lbx2
|
ladybird homeobox 2 |
| chr10_-_34058331 | 0.35 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr19_+_32570656 | 0.35 |
ENSDART00000005255
|
mrpl53
|
mitochondrial ribosomal protein L53 |
| chr10_-_25246786 | 0.34 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr9_+_28329384 | 0.34 |
ENSDART00000046880
|
plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
| KN149830v1_-_31870 | 0.34 |
ENSDART00000167653
|
zgc:194336
|
zgc:194336 |
| chr9_-_1701626 | 0.34 |
ENSDART00000163482
|
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr6_-_33893933 | 0.34 |
ENSDART00000129916
|
tmem69
|
transmembrane protein 69 |
| chr24_-_32259086 | 0.34 |
ENSDART00000048633
|
trdmt1
|
tRNA aspartic acid methyltransferase 1 |
| chr8_+_17739521 | 0.33 |
ENSDART00000112356
|
si:ch211-150o23.3
|
si:ch211-150o23.3 |
| chr13_-_9509729 | 0.33 |
ENSDART00000101949
|
sfxn4
|
sideroflexin 4 |
| chr14_-_14353487 | 0.32 |
ENSDART00000172241
|
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr8_-_11286377 | 0.32 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr4_+_74845382 | 0.32 |
ENSDART00000159789
|
zgc:172128
|
zgc:172128 |
| chr21_+_21158761 | 0.32 |
ENSDART00000058311
|
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr25_+_30699938 | 0.32 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr14_-_4038642 | 0.32 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr20_-_29961621 | 0.31 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr18_-_15582896 | 0.31 |
ENSDART00000172690
ENSDART00000159915 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr4_-_75732882 | 0.31 |
|
|
|
| chr21_+_27477153 | 0.31 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr13_-_36672285 | 0.31 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr2_-_17721810 | 0.30 |
ENSDART00000100201
ENSDART00000141188 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr10_+_33449922 | 0.30 |
ENSDART00000115379
ENSDART00000163458 ENSDART00000078012 |
zgc:153345
|
zgc:153345 |
| chr1_-_54570813 | 0.30 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr16_-_25452949 | 0.30 |
ENSDART00000167574
|
rbfa
|
ribosome binding factor A |
| chr15_+_34734212 | 0.30 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr9_-_22318918 | 0.30 |
ENSDART00000124272
|
crygm2d17
|
crystallin, gamma M2d17 |
| chr21_-_2350090 | 0.30 |
ENSDART00000168946
|
si:ch211-241b2.4
|
si:ch211-241b2.4 |
| chr11_+_17849608 | 0.30 |
ENSDART00000020283
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr20_-_29961589 | 0.29 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr3_-_6633512 | 0.28 |
ENSDART00000165273
|
GGA3 (1 of many)
|
golgi associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr20_+_34693415 | 0.28 |
ENSDART00000138338
|
ENSDARG00000054723
|
ENSDARG00000054723 |
| chr17_-_52233071 | 0.28 |
|
|
|
| chr3_-_15889742 | 0.28 |
ENSDART00000143324
|
spsb3a
|
splA/ryanodine receptor domain and SOCS box containing 3a |
| chr7_+_23752492 | 0.27 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr5_+_65633207 | 0.27 |
ENSDART00000114532
|
malt1
|
MALT paracaspase 1 |
| chr11_+_44108192 | 0.26 |
ENSDART00000165219
ENSDART00000160678 |
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr18_-_188494 | 0.26 |
ENSDART00000166207
ENSDART00000172235 ENSDART00000170884 |
gart
|
phosphoribosylglycinamide formyltransferase |
| chr21_+_25199691 | 0.26 |
ENSDART00000168140
ENSDART00000112783 |
tmem45b
|
transmembrane protein 45B |
| chr24_+_17115897 | 0.26 |
ENSDART00000129554
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr23_+_26806694 | 0.26 |
ENSDART00000035080
|
zgc:158263
|
zgc:158263 |
| chr21_+_25740782 | 0.26 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr24_-_8585312 | 0.26 |
|
|
|
| chr5_+_25133592 | 0.26 |
ENSDART00000098467
|
abhd17b
|
abhydrolase domain containing 17B |
| chr10_+_9575 | 0.26 |
|
|
|
| chr16_+_1227270 | 0.25 |
|
|
|
| chr4_+_5246465 | 0.25 |
ENSDART00000137966
|
ccdc167
|
coiled-coil domain containing 167 |
| chr10_-_25737632 | 0.25 |
ENSDART00000135058
|
sod1
|
superoxide dismutase 1, soluble |
| chr20_-_29961498 | 0.25 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr24_-_2278409 | 0.25 |
|
|
|
| chr5_+_51992974 | 0.25 |
ENSDART00000170341
|
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr8_+_23716843 | 0.25 |
ENSDART00000136547
|
rpl10a
|
ribosomal protein L10a |
| chr23_-_19299279 | 0.25 |
ENSDART00000080099
|
oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr2_+_47827350 | 0.24 |
|
|
|
| chr10_-_43874646 | 0.24 |
ENSDART00000025366
|
cetn3
|
centrin 3 |
| chr3_+_40022244 | 0.24 |
ENSDART00000102526
|
gfer
|
growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) |
| chr23_-_36922175 | 0.24 |
|
|
|
| chr7_+_21006803 | 0.24 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr18_+_1353738 | 0.24 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr2_+_21267793 | 0.24 |
ENSDART00000099913
|
pla2g4aa
|
phospholipase A2, group IVAa (cytosolic, calcium-dependent) |
| chr20_-_9107294 | 0.24 |
ENSDART00000140792
|
OMA1
|
OMA1 zinc metallopeptidase |
| chr17_-_9794176 | 0.24 |
|
|
|
| chr6_+_18941186 | 0.24 |
ENSDART00000044519
|
cx44.2
|
connexin 44.2 |
| chr16_+_3090170 | 0.24 |
ENSDART00000110395
|
limd1a
|
LIM domains containing 1a |
| chr15_+_9351511 | 0.23 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr3_+_28808901 | 0.23 |
ENSDART00000141904
ENSDART00000077221 |
lgals1l1
|
lectin, galactoside-binding, soluble, 1 (galectin 1)-like 1 |
| chr18_+_8943793 | 0.23 |
ENSDART00000144247
|
si:dkey-5i3.5
|
si:dkey-5i3.5 |
| chr9_+_24255064 | 0.23 |
ENSDART00000101577
ENSDART00000159324 ENSDART00000023196 ENSDART00000079689 ENSDART00000172743 ENSDART00000171577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr8_+_28493742 | 0.23 |
ENSDART00000053782
|
scrt2
|
scratch family zinc finger 2 |
| chr3_+_39437497 | 0.23 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr15_-_21003820 | 0.23 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr1_-_25272318 | 0.23 |
ENSDART00000134192
|
synpo2b
|
synaptopodin 2b |
| chr15_-_5827067 | 0.23 |
ENSDART00000102459
|
rbp2a
|
retinol binding protein 2a, cellular |
| chr13_+_33138144 | 0.22 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr16_+_53602834 | 0.22 |
ENSDART00000074653
|
grinab
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1b (glutamate binding) |
| chr19_+_12995955 | 0.22 |
ENSDART00000132892
|
cthrc1a
|
collagen triple helix repeat containing 1a |
| chr23_-_42775849 | 0.22 |
ENSDART00000149944
|
gpx7
|
glutathione peroxidase 7 |
| chr10_-_35293024 | 0.22 |
ENSDART00000145804
|
ypel2a
|
yippee-like 2a |
| chr2_-_49114158 | 0.22 |
|
|
|
| chr20_+_47639141 | 0.22 |
ENSDART00000043938
|
tram2
|
translocation associated membrane protein 2 |
| chr10_+_1653720 | 0.22 |
ENSDART00000018532
|
triap1
|
TP53 regulated inhibitor of apoptosis 1 |
| chr7_+_25052687 | 0.22 |
ENSDART00000110347
|
cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr19_+_44413865 | 0.22 |
ENSDART00000136147
ENSDART00000151004 |
nkd3
|
naked cuticle homolog 3 |
| chr11_+_5522377 | 0.21 |
ENSDART00000013203
|
cse1l
|
CSE1 chromosome segregation 1-like (yeast) |
| chr1_+_9633924 | 0.21 |
ENSDART00000029774
|
tmem55bb
|
transmembrane protein 55Bb |
| chr16_+_42567707 | 0.21 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr3_-_34006846 | 0.21 |
ENSDART00000136900
|
clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr15_-_16241341 | 0.21 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr23_-_32043229 | 0.21 |
ENSDART00000134550
|
ormdl2
|
ORMDL sphingolipid biosynthesis regulator 2 |
| chr18_+_21419735 | 0.21 |
ENSDART00000144523
|
necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr15_-_3023693 | 0.21 |
|
|
|
| chr20_-_27193158 | 0.20 |
ENSDART00000062096
|
si:dkeyp-55f12.3
|
si:dkeyp-55f12.3 |
| chr9_-_30765708 | 0.20 |
ENSDART00000101085
|
morc3a
|
MORC family CW-type zinc finger 3a |
| chr18_+_8362265 | 0.20 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr8_+_7740132 | 0.20 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr14_+_8634518 | 0.20 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr16_-_28723722 | 0.20 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr16_-_43061368 | 0.20 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr8_+_19945528 | 0.20 |
ENSDART00000134124
|
znf692
|
zinc finger protein 692 |
| KN150200v1_+_7856 | 0.20 |
|
|
|
| chr1_+_51087450 | 0.20 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr2_-_2218945 | 0.20 |
ENSDART00000156951
|
FP101911.1
|
ENSDARG00000097328 |
| chr5_+_31475897 | 0.20 |
ENSDART00000144510
|
zmat5
|
zinc finger, matrin-type 5 |
| chr16_-_28723759 | 0.19 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr14_+_51934755 | 0.19 |
|
|
|
| chr3_+_479366 | 0.19 |
|
|
|
| chr14_-_8634381 | 0.19 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
| chr7_+_23752651 | 0.19 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr23_-_23474703 | 0.19 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr1_-_9256864 | 0.19 |
ENSDART00000041849
|
tmem8a
|
transmembrane protein 8A |
| chr4_-_951970 | 0.19 |
ENSDART00000093289
|
xrcc6bp1
|
XRCC6 binding protein 1 |
| chr2_-_58111727 | 0.19 |
ENSDART00000004431
ENSDART00000166845 ENSDART00000163999 |
epb41l3b
|
erythrocyte membrane protein band 4.1-like 3b |
| chr19_-_22895095 | 0.19 |
|
|
|
| chr7_-_73866202 | 0.19 |
|
|
|
| chr14_-_45883839 | 0.19 |
|
|
|
| chr2_+_31974269 | 0.19 |
ENSDART00000012413
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr19_+_44185325 | 0.19 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr5_-_40894631 | 0.19 |
ENSDART00000121840
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr21_+_22598805 | 0.19 |
ENSDART00000142495
|
BX510940.1
|
ENSDARG00000089088 |
| chr14_-_50075366 | 0.19 |
ENSDART00000021260
|
sept8b
|
septin 8b |
| chr2_+_57734025 | 0.19 |
ENSDART00000143453
|
plekhj1
|
pleckstrin homology domain containing, family J member 1 |
| chr3_-_21149752 | 0.18 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr6_-_7577736 | 0.18 |
ENSDART00000151545
|
slc25a38b
|
solute carrier family 25, member 38b |
| chr15_+_35076414 | 0.18 |
ENSDART00000165210
|
zgc:66024
|
zgc:66024 |
| chr23_+_11412329 | 0.18 |
ENSDART00000135406
|
chl1a
|
cell adhesion molecule L1-like a |
| chr23_+_36023748 | 0.18 |
|
|
|
| chr14_-_30630835 | 0.18 |
ENSDART00000010512
|
zgc:92907
|
zgc:92907 |
| chr24_-_10935904 | 0.18 |
ENSDART00000003195
|
chmp4c
|
charged multivesicular body protein 4C |
| chr22_+_16509286 | 0.18 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr19_-_18664670 | 0.18 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr18_+_14715573 | 0.18 |
ENSDART00000108469
|
spata2l
|
spermatogenesis associated 2-like |
| chr16_+_34038263 | 0.18 |
ENSDART00000134946
|
zdhhc18a
|
zinc finger, DHHC-type containing 18a |
| chr21_+_28921734 | 0.18 |
ENSDART00000166575
|
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr9_-_43736549 | 0.18 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr11_-_38856849 | 0.18 |
ENSDART00000113185
|
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr13_+_40347099 | 0.18 |
|
|
|
| chr14_-_17257773 | 0.18 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor-like 1a |
| chr19_-_35739239 | 0.18 |
|
|
|
| chr17_-_21398340 | 0.18 |
ENSDART00000007021
|
atp6v1ba
|
ATPase, H+ transporting, lysosomal, V1 subunit B, member a |
| chr2_+_11902170 | 0.18 |
ENSDART00000138562
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr22_-_19986461 | 0.17 |
ENSDART00000093310
|
celf5a
|
cugbp, Elav-like family member 5a |
| chr4_+_22759177 | 0.17 |
ENSDART00000146272
|
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr3_+_26158051 | 0.17 |
ENSDART00000163500
|
rhot1a
|
ras homolog family member T1a |
| chr22_+_11943032 | 0.17 |
ENSDART00000105788
|
mgat5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr11_+_35790782 | 0.17 |
ENSDART00000125221
|
CR933559.1
|
ENSDARG00000087939 |
| chr3_-_48458042 | 0.17 |
ENSDART00000156822
|
cdip1
|
cell death-inducing p53 target 1 |
| chr20_-_47994364 | 0.17 |
ENSDART00000166857
|
CABZ01059120.1
|
ENSDARG00000100162 |
| chr20_+_19613133 | 0.17 |
ENSDART00000152548
ENSDART00000063696 |
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr9_+_36092350 | 0.17 |
ENSDART00000005086
|
atp1a1b
|
ATPase, Na+/K+ transporting, alpha 1b polypeptide |
| chr25_+_4534280 | 0.17 |
ENSDART00000130299
|
deaf1
|
DEAF1 transcription factor |
| chr21_-_13026077 | 0.17 |
ENSDART00000024616
|
fam219aa
|
family with sequence similarity 219, member Aa |
| chr20_+_1724609 | 0.17 |
|
|
|
| chr9_+_20715436 | 0.17 |
ENSDART00000141775
|
trim45
|
tripartite motif containing 45 |
| chr11_+_15935769 | 0.17 |
ENSDART00000158824
|
ctbs
|
chitobiase, di-N-acetyl- |
| chr22_-_19986607 | 0.17 |
ENSDART00000093310
|
celf5a
|
cugbp, Elav-like family member 5a |
| KN150037v1_+_2317 | 0.16 |
|
|
|
| chr21_+_31113731 | 0.16 |
ENSDART00000065366
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr1_+_51706536 | 0.16 |
|
|
|
| chr24_+_8702288 | 0.16 |
ENSDART00000114810
|
sycp2l
|
synaptonemal complex protein 2-like |
| chr5_-_31796734 | 0.16 |
ENSDART00000142095
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr15_-_5636737 | 0.16 |
ENSDART00000114410
|
wdr62
|
WD repeat domain 62 |
| chr2_+_51087570 | 0.16 |
|
|
|
| chr13_-_24778226 | 0.16 |
|
|
|
| chr19_+_5562107 | 0.16 |
ENSDART00000082080
|
jupb
|
junction plakoglobin b |
| chr12_-_7902815 | 0.16 |
ENSDART00000088100
|
ank3b
|
ankyrin 3b |
| chr8_-_12253026 | 0.16 |
ENSDART00000091612
|
dab2ipa
|
DAB2 interacting protein a |
| chr9_-_30553051 | 0.16 |
ENSDART00000147030
|
sytl5
|
synaptotagmin-like 5 |
| chr6_+_8362892 | 0.16 |
ENSDART00000032118
|
ube2g2
|
ubiquitin-conjugating enzyme E2G 2 (UBC7 homolog, yeast) |
| chr17_+_24300332 | 0.15 |
ENSDART00000064083
|
otx1b
|
orthodenticle homeobox 1b |
| chr19_-_5186692 | 0.15 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr21_-_2273243 | 0.15 |
ENSDART00000168712
|
si:ch73-299h12.6
|
si:ch73-299h12.6 |
| chr22_+_35229139 | 0.15 |
ENSDART00000061315
ENSDART00000146430 |
tsc22d2
|
TSC22 domain family 2 |
| chr19_-_22182031 | 0.15 |
ENSDART00000104279
|
znf516
|
zinc finger protein 516 |
| chr19_-_2084281 | 0.15 |
|
|
|
| chr6_+_18941289 | 0.15 |
ENSDART00000044519
|
cx44.2
|
connexin 44.2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:1901909 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.4 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.1 | 0.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.4 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.3 | GO:0070658 | mechanosensory epithelium regeneration(GO:0070655) mechanoreceptor differentiation involved in mechanosensory epithelium regeneration(GO:0070656) neuromast regeneration(GO:0070657) neuromast hair cell differentiation involved in neuromast regeneration(GO:0070658) |
| 0.1 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.2 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.1 | 0.2 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.4 | GO:0046084 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.1 | 0.4 | GO:1990748 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 0.2 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 0.2 | GO:0090594 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) inflammatory response to wounding(GO:0090594) |
| 0.1 | 0.4 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 0.5 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.2 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.2 | GO:0045669 | regulation of osteoblast proliferation(GO:0033688) positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.3 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.0 | 0.3 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.6 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.2 | GO:0007589 | body fluid secretion(GO:0007589) |
| 0.0 | 0.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.2 | GO:0060148 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.2 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.0 | 0.1 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.0 | 0.4 | GO:0006555 | methionine metabolic process(GO:0006555) methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.3 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.2 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.2 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.1 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.0 | 0.3 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0042423 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.0 | 0.3 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.4 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.1 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.1 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.2 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:0007168 | cGMP biosynthetic process(GO:0006182) receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.8 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.3 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.2 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.1 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 0.1 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.0 | 0.2 | GO:0051654 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) establishment of mitochondrion localization(GO:0051654) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.0 | 0.3 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.4 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.3 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.3 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.2 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.3 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.0 | GO:0019478 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.0 | 0.1 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 0.5 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.1 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.3 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.1 | GO:1900153 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.2 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.3 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.4 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.6 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.1 | 0.9 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.4 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.3 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.1 | 0.3 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.1 | 0.3 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 0.1 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.4 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.2 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.2 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.2 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.3 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.3 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.1 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 0.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.3 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.3 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.5 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.0 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.2 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.0 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.0 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.5 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |