Project
DANIO-CODE
Navigation
Downloads
Results for ar_nr3c1
Z-value: 2.04
Transcription factors associated with ar_nr3c1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
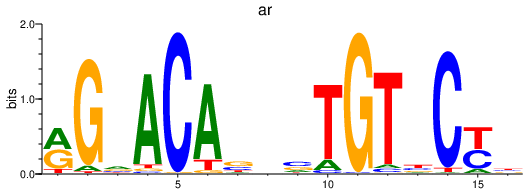
ar
|
ENSDARG00000067976 | androgen receptor |
|
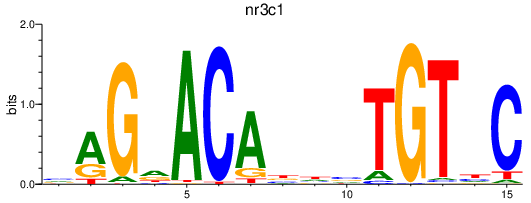
nr3c1
|
ENSDARG00000025032 | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr3c1 | dr10_dc_chr14_-_23503974_23504137 | 0.68 | 3.5e-03 | Click! |
Activity profile of ar_nr3c1 motif
Sorted Z-values of ar_nr3c1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ar_nr3c1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_45258675 | 7.29 |
ENSDART00000158855
|
fxyd1
|
FXYD domain containing ion transport regulator 1 (phospholemman) |
| chr15_-_9055873 | 7.25 |
ENSDART00000124998
|
rtn2a
|
reticulon 2a |
| chr21_+_11410738 | 6.84 |
ENSDART00000146701
|
si:dkey-184p9.7
|
si:dkey-184p9.7 |
| chr22_-_951880 | 6.76 |
ENSDART00000105895
ENSDART00000172206 |
cacna1sa
|
calcium channel, voltage-dependent, L type, alpha 1S subunit, a |
| chr19_+_10936229 | 5.90 |
ENSDART00000172219
|
apoea
|
apolipoprotein Ea |
| KN149955v1_+_4206 | 5.83 |
ENSDART00000167370
|
cdkn1d
|
cyclin-dependent kinase inhibitor 1D |
| chr13_+_39110160 | 5.59 |
ENSDART00000111458
|
zgc:172136
|
zgc:172136 |
| chr16_+_32605735 | 5.49 |
ENSDART00000093250
|
pou3f2b
|
POU class 3 homeobox 2b |
| chr10_+_26705729 | 5.34 |
ENSDART00000147013
|
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr19_+_7798512 | 5.24 |
ENSDART00000104719
ENSDART00000146747 |
tuft1b
|
tuftelin 1b |
| chr5_-_43334777 | 5.16 |
ENSDART00000142271
|
ENSDARG00000053091
|
ENSDARG00000053091 |
| chr7_+_58397256 | 5.11 |
ENSDART00000049264
|
sdr16c5b
|
short chain dehydrogenase/reductase family 16C, member 5b |
| chr18_-_39491932 | 4.80 |
ENSDART00000122930
|
scg3
|
secretogranin III |
| chr15_-_23407388 | 4.52 |
ENSDART00000020425
|
mcamb
|
melanoma cell adhesion molecule b |
| chr11_+_13470883 | 4.28 |
ENSDART00000011362
|
arrdc2
|
arrestin domain containing 2 |
| chr4_+_4795126 | 4.21 |
ENSDART00000129252
|
slc13a4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr13_+_28689749 | 4.21 |
ENSDART00000101653
|
CU639469.1
|
ENSDARG00000062790 |
| chr25_+_3232912 | 4.09 |
ENSDART00000104877
ENSDART00000170765 ENSDART00000158571 |
chchd3b
|
coiled-coil-helix-coiled-coil-helix domain containing 3b |
| chr16_+_10885785 | 3.98 |
ENSDART00000161969
|
atp1a3b
|
ATPase, Na+/K+ transporting, alpha 3b polypeptide |
| chr11_-_25592102 | 3.89 |
ENSDART00000103549
|
skib
|
v-ski avian sarcoma viral oncogene homolog b |
| chr18_-_48750637 | 3.77 |
ENSDART00000097259
|
st3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr18_-_25010085 | 3.66 |
ENSDART00000133786
|
si:ch211-196l7.4
|
si:ch211-196l7.4 |
| chr9_+_49054601 | 3.59 |
ENSDART00000047401
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr21_-_25528699 | 3.55 |
ENSDART00000144917
|
ENSDARG00000089836
|
ENSDARG00000089836 |
| chr5_+_31606213 | 3.53 |
ENSDART00000126873
|
myhz2
|
myosin, heavy polypeptide 2, fast muscle specific |
| chr20_+_37940880 | 3.50 |
ENSDART00000165887
|
flvcr1
|
feline leukemia virus subgroup C cellular receptor 1 |
| chr11_-_25592017 | 3.34 |
ENSDART00000103549
|
skib
|
v-ski avian sarcoma viral oncogene homolog b |
| chr23_+_35609887 | 3.28 |
ENSDART00000179393
|
tuba1b
|
tubulin, alpha 1b |
| chr8_+_7340538 | 3.21 |
ENSDART00000121708
|
pcsk1nl
|
proprotein convertase subtilisin/kexin type 1 inhibitor, like |
| chr16_-_32605560 | 3.20 |
|
|
|
| chr8_-_4903896 | 3.16 |
ENSDART00000004986
|
igsf9b
|
immunoglobulin superfamily, member 9b |
| chr4_+_4223606 | 3.10 |
|
|
|
| chr1_+_23385409 | 2.98 |
ENSDART00000014608
|
mab21l2
|
mab-21-like 2 |
| chr23_+_37355971 | 2.96 |
|
|
|
| chr3_+_23563620 | 2.94 |
ENSDART00000147022
|
hoxb7a
|
homeobox B7a |
| chr5_+_22752368 | 2.90 |
ENSDART00000142915
|
si:dkey-125i10.3
|
si:dkey-125i10.3 |
| chr20_+_25441689 | 2.85 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr4_-_12726432 | 2.84 |
ENSDART00000132286
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr20_-_6822668 | 2.82 |
ENSDART00000169569
|
igfbp1a
|
insulin-like growth factor binding protein 1a |
| chr2_-_48341913 | 2.81 |
ENSDART00000139944
|
si:dkeyp-12a9.5
|
si:dkeyp-12a9.5 |
| chr25_-_20994084 | 2.75 |
ENSDART00000154765
|
prr5a
|
proline rich 5a (renal) |
| chr17_+_29328653 | 2.74 |
ENSDART00000086164
|
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr12_-_46943791 | 2.74 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr24_+_35676505 | 2.70 |
ENSDART00000122734
|
cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr3_-_54866453 | 2.69 |
ENSDART00000125092
|
hbae1
|
hemoglobin, alpha embryonic 1 |
| chr9_-_47042015 | 2.64 |
ENSDART00000054137
|
igfbp5b
|
insulin-like growth factor binding protein 5b |
| chr18_+_26437604 | 2.62 |
ENSDART00000060245
|
ctsh
|
cathepsin H |
| chr1_+_30941618 | 2.57 |
ENSDART00000044214
|
wbp1lb
|
WW domain binding protein 1-like b |
| chr14_-_27703812 | 2.54 |
ENSDART00000054115
|
tsc22d3
|
TSC22 domain family, member 3 |
| chr21_-_19280266 | 2.53 |
ENSDART00000141596
|
gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr23_+_45020761 | 2.51 |
ENSDART00000159104
|
atp1b2a
|
ATPase, Na+/K+ transporting, beta 2a polypeptide |
| chr18_-_35755677 | 2.50 |
ENSDART00000036015
|
ryr1b
|
ryanodine receptor 1b (skeletal) |
| KN149955v1_+_4134 | 2.48 |
ENSDART00000167370
|
cdkn1d
|
cyclin-dependent kinase inhibitor 1D |
| chr9_+_1225808 | 2.45 |
ENSDART00000177730
|
CABZ01114359.1
|
ENSDARG00000107255 |
| chr18_-_48750433 | 2.44 |
ENSDART00000097259
|
st3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr23_-_39956151 | 2.43 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr21_+_28441951 | 2.32 |
ENSDART00000077887
|
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr19_+_17926983 | 2.31 |
ENSDART00000141397
|
ube2e1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr12_-_28456044 | 2.30 |
ENSDART00000008010
|
pdk2a
|
pyruvate dehydrogenase kinase, isozyme 2a |
| chr3_-_30948297 | 2.30 |
ENSDART00000145636
|
ELOB (1 of many)
|
elongin B |
| chr15_-_29454583 | 2.28 |
ENSDART00000145976
|
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr9_-_30437160 | 2.28 |
ENSDART00000147241
|
BX936337.1
|
ENSDARG00000092870 |
| chr3_-_35671276 | 2.26 |
ENSDART00000150911
|
caskin1
|
CASK interacting protein 1 |
| chr16_+_330953 | 2.26 |
ENSDART00000102977
|
nrsn1
|
neurensin 1 |
| chr18_+_17439409 | 2.23 |
ENSDART00000100223
|
zgc:91860
|
zgc:91860 |
| chr4_-_20435099 | 2.17 |
ENSDART00000055317
|
lrrc17
|
leucine rich repeat containing 17 |
| chr20_+_34640435 | 2.14 |
ENSDART00000152073
|
ENSDARG00000054723
|
ENSDARG00000054723 |
| chr16_-_46463900 | 2.10 |
ENSDART00000157045
|
CU861453.1
|
ENSDARG00000097192 |
| chr19_-_41819752 | 2.08 |
ENSDART00000167772
|
shfm1
|
split hand/foot malformation (ectrodactyly) type 1 |
| chr24_+_7607915 | 2.04 |
ENSDART00000124409
|
cavin1b
|
caveolae associated protein 1b |
| chr23_-_20332076 | 2.03 |
ENSDART00000147326
|
lamb2
|
laminin, beta 2 (laminin S) |
| chr3_-_58488929 | 2.03 |
ENSDART00000042386
|
unm_sa1261
|
un-named sa1261 |
| chr16_-_46463845 | 2.03 |
ENSDART00000157045
|
CU861453.1
|
ENSDARG00000097192 |
| chr1_-_32934338 | 2.02 |
|
|
|
| chr2_+_52639037 | 2.01 |
ENSDART00000006216
|
plpp2a
|
phospholipid phosphatase 2a |
| chr8_+_19642272 | 1.96 |
ENSDART00000138176
|
foxd2
|
forkhead box D2 |
| chr10_-_43466392 | 1.95 |
ENSDART00000062631
|
hapln1b
|
hyaluronan and proteoglycan link protein 1b |
| chr16_-_31835463 | 1.95 |
ENSDART00000148389
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr9_+_38975508 | 1.94 |
|
|
|
| chr22_-_13325409 | 1.93 |
ENSDART00000154095
|
si:ch211-227m13.1
|
si:ch211-227m13.1 |
| chr18_+_27119086 | 1.92 |
ENSDART00000086094
|
plekha7a
|
pleckstrin homology domain containing, family A member 7a |
| chr12_-_4648262 | 1.90 |
ENSDART00000152771
|
si:ch211-255p10.3
|
si:ch211-255p10.3 |
| chr2_+_13373937 | 1.89 |
ENSDART00000142649
|
prkag2b
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit b |
| chr17_+_6119323 | 1.87 |
ENSDART00000131075
|
dusp23b
|
dual specificity phosphatase 23b |
| chr5_-_40894631 | 1.85 |
ENSDART00000121840
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr10_-_43466478 | 1.85 |
ENSDART00000062631
|
hapln1b
|
hyaluronan and proteoglycan link protein 1b |
| chr16_-_17439735 | 1.85 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr2_-_24113073 | 1.84 |
ENSDART00000148685
|
xirp1
|
xin actin binding repeat containing 1 |
| chr25_+_4918339 | 1.84 |
ENSDART00000153980
|
parvb
|
parvin, beta |
| chr7_-_7442720 | 1.83 |
ENSDART00000091105
|
mfap3l
|
microfibrillar-associated protein 3-like |
| chr5_-_31301280 | 1.80 |
ENSDART00000141446
|
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr20_+_230394 | 1.79 |
ENSDART00000002661
|
lama4
|
laminin, alpha 4 |
| chr17_+_52736192 | 1.78 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr3_+_16115708 | 1.77 |
ENSDART00000122519
|
st8sia6
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
| chr1_+_4557485 | 1.77 |
ENSDART00000052423
|
spry2
|
sprouty RTK signaling antagonist 2 |
| chr19_+_31046291 | 1.74 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr19_+_19839981 | 1.73 |
ENSDART00000113580
|
ENSDARG00000074216
|
ENSDARG00000074216 |
| chr12_-_16331781 | 1.68 |
ENSDART00000152133
|
hectd2
|
HECT domain containing 2 |
| chr16_-_21238288 | 1.67 |
ENSDART00000139737
|
cbx3b
|
chromobox homolog 3b |
| chr6_-_49064365 | 1.67 |
ENSDART00000156124
|
si:ch211-105j21.9
|
si:ch211-105j21.9 |
| chr20_+_2255204 | 1.66 |
ENSDART00000137579
|
si:ch73-18b11.2
|
si:ch73-18b11.2 |
| chr1_-_53247258 | 1.66 |
ENSDART00000007732
|
capn9
|
calpain 9 |
| chr6_+_8895437 | 1.64 |
ENSDART00000162588
|
rgn
|
regucalcin |
| chr3_-_32205873 | 1.62 |
ENSDART00000156918
|
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr23_-_29138952 | 1.61 |
ENSDART00000002812
|
casz1
|
castor zinc finger 1 |
| chr5_-_28888704 | 1.55 |
|
|
|
| chr13_+_22119569 | 1.54 |
ENSDART00000108472
|
synpo2la
|
synaptopodin 2-like a |
| chr17_-_37447917 | 1.54 |
ENSDART00000075975
|
crip1
|
cysteine-rich protein 1 |
| chr13_-_1177857 | 1.54 |
ENSDART00000111052
|
olig3
|
oligodendrocyte transcription factor 3 |
| chr1_-_44348236 | 1.53 |
ENSDART00000023336
|
rps6
|
ribosomal protein S6 |
| chr4_-_16417703 | 1.51 |
ENSDART00000013085
|
dcn
|
decorin |
| chr14_+_20632593 | 1.51 |
ENSDART00000166366
ENSDART00000106198 |
zgc:66433
|
zgc:66433 |
| chr2_+_26273986 | 1.50 |
ENSDART00000137746
|
slc7a14a
|
solute carrier family 7, member 14a |
| chr17_+_381715 | 1.50 |
ENSDART00000162898
|
si:rp71-62i8.1
|
si:rp71-62i8.1 |
| chr18_+_16755239 | 1.50 |
ENSDART00000133490
|
lyve1b
|
lymphatic vessel endothelial hyaluronic receptor 1b |
| chr17_+_52024610 | 1.48 |
|
|
|
| chr23_-_5751064 | 1.47 |
ENSDART00000067351
|
tnnt2a
|
troponin T type 2a (cardiac) |
| chr23_-_28367816 | 1.46 |
ENSDART00000003548
|
znf385a
|
zinc finger protein 385A |
| chr5_-_69628472 | 1.45 |
|
|
|
| chr2_+_3701942 | 1.44 |
ENSDART00000132572
|
gpt2l
|
glutamic pyruvate transaminase (alanine aminotransferase) 2, like |
| chr16_-_12375204 | 1.44 |
ENSDART00000113345
|
leng8
|
leukocyte receptor cluster (LRC) member 8 |
| chr10_-_36864268 | 1.43 |
ENSDART00000165853
|
dhrs13a.1
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 |
| chr18_+_45121016 | 1.43 |
ENSDART00000172328
|
gyltl1b
|
glycosyltransferase-like 1b |
| chr15_+_161444 | 1.42 |
ENSDART00000008504
|
med17
|
mediator complex subunit 17 |
| chr9_-_32942783 | 1.42 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr10_-_37131530 | 1.40 |
ENSDART00000132023
|
myo18aa
|
myosin XVIIIAa |
| chr25_-_18234069 | 1.39 |
ENSDART00000104496
|
dusp6
|
dual specificity phosphatase 6 |
| chr1_+_15827083 | 1.39 |
|
|
|
| chr20_-_46458839 | 1.36 |
ENSDART00000153087
|
bmf2
|
Bcl2 modifying factor 2 |
| chr17_+_52736535 | 1.35 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr4_-_2595775 | 1.35 |
ENSDART00000132581
ENSDART00000019508 |
csrp2
|
cysteine and glycine-rich protein 2 |
| chr11_-_53846 | 1.34 |
|
|
|
| chr1_-_58242892 | 1.34 |
|
|
|
| chr23_+_17939650 | 1.33 |
ENSDART00000162822
|
naca
|
nascent polypeptide-associated complex alpha subunit |
| chr12_+_18643598 | 1.32 |
|
|
|
| chr11_+_132668 | 1.31 |
ENSDART00000163978
ENSDART00000170778 |
dusp7
|
dual specificity phosphatase 7 |
| chr19_+_44185325 | 1.31 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr5_-_28888658 | 1.28 |
|
|
|
| chr7_-_23601966 | 1.26 |
ENSDART00000101408
|
zgc:162171
|
zgc:162171 |
| chr8_+_999710 | 1.26 |
ENSDART00000081432
|
sprb
|
sepiapterin reductase b |
| chr24_-_20812255 | 1.25 |
ENSDART00000142080
|
kpna1
|
karyopherin alpha 1 (importin alpha 5) |
| chr15_+_15479719 | 1.25 |
ENSDART00000127436
|
zgc:92630
|
zgc:92630 |
| chr20_-_34898276 | 1.23 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr25_-_23949331 | 1.23 |
ENSDART00000047569
|
igf2b
|
insulin-like growth factor 2b |
| chr18_+_29178482 | 1.23 |
ENSDART00000137587
ENSDART00000144423 |
ppfibp2a
|
PTPRF interacting protein, binding protein 2a (liprin beta 2) |
| chr23_+_4955111 | 1.20 |
ENSDART00000023537
|
tnnc1a
|
troponin C type 1a (slow) |
| chr4_-_72067910 | 1.19 |
ENSDART00000170308
|
BX855614.3
|
ENSDARG00000101193 |
| chr17_-_45403518 | 1.19 |
ENSDART00000156002
|
tmem206
|
transmembrane protein 206 |
| chr5_+_50947650 | 1.18 |
ENSDART00000058403
|
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr4_+_4795530 | 1.17 |
ENSDART00000129252
|
slc13a4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr19_-_31785558 | 1.16 |
ENSDART00000046609
|
scin
|
scinderin |
| chr14_+_29428950 | 1.16 |
ENSDART00000105898
|
ENSDARG00000071567
|
ENSDARG00000071567 |
| chr3_-_33290990 | 1.16 |
ENSDART00000075493
ENSDART00000132293 |
ccdc103
|
coiled-coil domain containing 103 |
| chr19_+_5501487 | 1.16 |
ENSDART00000132874
|
eif1b
|
eukaryotic translation initiation factor 1B |
| chr22_-_13325475 | 1.15 |
ENSDART00000154095
|
si:ch211-227m13.1
|
si:ch211-227m13.1 |
| chr12_-_26291790 | 1.15 |
ENSDART00000152941
|
usp54b
|
ubiquitin specific peptidase 54b |
| chr1_+_33510660 | 1.13 |
ENSDART00000147201
|
slc5a7a
|
solute carrier family 5 (sodium/choline cotransporter), member 7a |
| chr1_-_54319671 | 1.12 |
ENSDART00000074058
|
slc25a23a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23a |
| chr12_-_43939028 | 1.11 |
ENSDART00000170723
|
zgc:112964
|
zgc:112964 |
| chr19_-_3547521 | 1.11 |
ENSDART00000168139
|
hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr17_-_52735615 | 1.09 |
|
|
|
| chr4_+_27141061 | 1.08 |
ENSDART00000145083
|
brd1a
|
bromodomain containing 1a |
| chr25_-_20993989 | 1.07 |
|
|
|
| chr25_+_446920 | 1.07 |
ENSDART00000104717
|
rsl24d1
|
ribosomal L24 domain containing 1 |
| chr9_-_38232999 | 1.07 |
ENSDART00000133998
|
hacd2
|
3-hydroxyacyl-CoA dehydratase 2 |
| chr21_+_25017684 | 1.05 |
ENSDART00000065132
|
zgc:171740
|
zgc:171740 |
| chr3_-_54414513 | 1.04 |
ENSDART00000053106
|
s1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr13_-_219661 | 1.03 |
ENSDART00000133731
|
si:ch1073-291c23.2
|
si:ch1073-291c23.2 |
| chr4_-_16365281 | 1.02 |
ENSDART00000139919
|
lum
|
lumican |
| chr4_-_73484546 | 1.01 |
|
|
|
| chr4_+_19545842 | 1.01 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr11_-_23333850 | 0.98 |
|
|
|
| chr21_+_11046459 | 0.97 |
ENSDART00000169024
|
agxt2
|
alanine--glyoxylate aminotransferase 2 |
| chr20_+_46683031 | 0.96 |
ENSDART00000153231
|
jdp2b
|
Jun dimerization protein 2b |
| chr14_-_9216303 | 0.95 |
ENSDART00000054689
|
atoh8
|
atonal bHLH transcription factor 8 |
| chr24_-_32750010 | 0.94 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr9_+_30774173 | 0.94 |
ENSDART00000160590
|
tbc1d4
|
TBC1 domain family, member 4 |
| chr9_-_43736549 | 0.94 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr14_+_51692872 | 0.93 |
ENSDART00000163856
|
noa1
|
nitric oxide associated 1 |
| chr2_-_3571821 | 0.92 |
ENSDART00000092741
|
snap47
|
synaptosomal-associated protein, 47 |
| chr3_+_19863136 | 0.92 |
ENSDART00000078974
|
tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr24_-_21750082 | 0.91 |
|
|
|
| chr14_-_12001505 | 0.91 |
ENSDART00000171136
|
myot
|
myotilin |
| chr6_+_23462412 | 0.89 |
ENSDART00000166192
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr16_-_5936637 | 0.88 |
ENSDART00000085678
|
trak1
|
trafficking protein, kinesin binding 1 |
| chr16_-_46653204 | 0.88 |
ENSDART00000158341
|
tmem176l.3a
|
transmembrane protein 176l.3a |
| chr10_-_38372282 | 0.88 |
ENSDART00000149580
|
nrip1b
|
nuclear receptor interacting protein 1b |
| chr2_+_54911012 | 0.87 |
ENSDART00000027313
|
ndufv2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2 |
| chr19_-_9779467 | 0.87 |
|
|
|
| chr15_-_33782909 | 0.87 |
ENSDART00000167705
|
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr4_-_19992951 | 0.84 |
ENSDART00000169248
|
drd4-rs
|
dopamine receptor D4 related sequence |
| chr16_+_46463904 | 0.84 |
|
|
|
| chr10_-_13220838 | 0.84 |
ENSDART00000160265
|
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr24_+_21828704 | 0.84 |
ENSDART00000042495
|
sat1b
|
spermidine/spermine N1-acetyltransferase 1b |
| chr1_-_58258428 | 0.83 |
|
|
|
| chr14_-_2437593 | 0.83 |
ENSDART00000157677
|
si:ch73-233f7.6
|
si:ch73-233f7.6 |
| chr11_-_9479592 | 0.83 |
ENSDART00000175829
|
CU179643.1
|
ENSDARG00000086489 |
| chr5_-_40894693 | 0.83 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr21_-_19789339 | 0.82 |
|
|
|
| chr25_+_28329427 | 0.82 |
ENSDART00000160619
|
slc41a2a
|
solute carrier family 41 (magnesium transporter), member 2a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.9 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) positive regulation of nitric-oxide synthase activity(GO:0051000) beta-amyloid clearance(GO:0097242) regulation of beta-amyloid clearance(GO:1900221) |
| 1.4 | 5.6 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 1.2 | 3.5 | GO:0097037 | heme export(GO:0097037) |
| 0.7 | 2.2 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.6 | 1.9 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.6 | 2.5 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.6 | 3.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.6 | 5.8 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.5 | 1.5 | GO:0007263 | atrial cardiac muscle tissue development(GO:0003228) nitric oxide mediated signal transduction(GO:0007263) |
| 0.5 | 1.4 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.5 | 1.4 | GO:1905178 | regulation of cardiac muscle tissue regeneration(GO:1905178) |
| 0.5 | 2.8 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.4 | 1.6 | GO:0050848 | L-ascorbic acid metabolic process(GO:0019852) water-soluble vitamin biosynthetic process(GO:0042364) regulation of calcium-mediated signaling(GO:0050848) |
| 0.4 | 6.5 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.4 | 3.0 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.3 | 3.8 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.3 | 1.0 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.3 | 0.9 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.3 | 2.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.3 | 6.3 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.3 | 0.8 | GO:0030815 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) negative regulation of purine nucleotide biosynthetic process(GO:1900372) |
| 0.3 | 1.0 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.3 | 1.8 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.3 | 4.5 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.2 | 4.7 | GO:0014823 | response to activity(GO:0014823) |
| 0.2 | 0.7 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.2 | 2.5 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.2 | 1.1 | GO:1900619 | neuromuscular synaptic transmission(GO:0007274) acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.2 | 1.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.2 | 0.8 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 | 0.6 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.2 | 1.8 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.2 | 0.5 | GO:0032877 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.2 | 7.4 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.2 | 1.5 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.2 | 0.8 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.2 | 5.5 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.2 | 0.6 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.2 | 1.4 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.2 | 3.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 0.5 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.1 | 1.2 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 1.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.5 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 1.2 | GO:0090660 | cerebrospinal fluid circulation(GO:0090660) |
| 0.1 | 1.3 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 6.8 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.1 | 3.5 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.1 | 0.4 | GO:1990092 | calcium-dependent self proteolysis(GO:1990092) |
| 0.1 | 6.4 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.1 | 1.2 | GO:0032946 | positive regulation of mononuclear cell proliferation(GO:0032946) positive regulation of T cell proliferation(GO:0042102) positive regulation of lymphocyte proliferation(GO:0050671) positive regulation of leukocyte proliferation(GO:0070665) |
| 0.1 | 0.3 | GO:0035552 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded DNA demethylation(GO:0035552) regulation of multicellular organism growth(GO:0040014) |
| 0.1 | 0.3 | GO:0015822 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.1 | 2.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 0.3 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.1 | 0.3 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 0.6 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 1.1 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.7 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.1 | 1.8 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 0.7 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.1 | 1.0 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 0.4 | GO:0003379 | establishment of cell polarity involved in gastrulation cell migration(GO:0003379) |
| 0.1 | 1.3 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 0.9 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 1.9 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 1.5 | GO:0050654 | chondroitin sulfate proteoglycan metabolic process(GO:0050654) |
| 0.1 | 3.2 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 0.7 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.1 | 5.4 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.1 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 1.5 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 1.1 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.5 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 2.1 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 1.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 2.7 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.1 | 2.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.9 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 1.7 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.6 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 1.4 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 1.2 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.6 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.7 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 1.7 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 2.2 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.6 | GO:1902667 | regulation of axon guidance(GO:1902667) |
| 0.0 | 0.9 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 3.8 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 1.0 | GO:1902749 | regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.0 | 0.6 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.1 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.0 | 1.3 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.8 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 3.1 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 1.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.8 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 1.0 | GO:0071560 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 1.0 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 1.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.4 | GO:0003146 | heart jogging(GO:0003146) |
| 0.0 | 0.5 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.1 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 1.2 | GO:0006413 | translational initiation(GO:0006413) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.9 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.4 | 1.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.4 | 4.8 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.4 | 2.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.3 | 3.8 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.3 | 1.9 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.3 | 2.5 | GO:1904949 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.3 | 1.2 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.3 | 6.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.3 | 3.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 0.6 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.2 | 1.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 3.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 2.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 1.2 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 5.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.9 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 0.6 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 6.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 1.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 2.9 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 5.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 4.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 1.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.3 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.9 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 3.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.2 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.2 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.0 | 1.3 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0005903 | brush border(GO:0005903) brush border membrane(GO:0031526) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 3.2 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.9 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.3 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 3.8 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 3.3 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.4 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.6 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.8 | 8.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.7 | 2.9 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.7 | 6.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.6 | 5.6 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.6 | 5.8 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.5 | 2.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.4 | 2.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.4 | 2.5 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.4 | 4.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.4 | 1.1 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.4 | 5.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 3.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 2.3 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.2 | 2.7 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 2.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 4.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 8.0 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.2 | 1.1 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.2 | 0.8 | GO:0099528 | G-protein coupled neurotransmitter receptor activity(GO:0099528) |
| 0.2 | 0.8 | GO:0019809 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.2 | 0.6 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.2 | 2.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 1.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 1.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 4.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 0.6 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.2 | 1.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 0.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 2.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.2 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 1.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 7.2 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 0.4 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.7 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 2.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.3 | GO:0035515 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 0.3 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 1.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.3 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.5 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.1 | 0.7 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 1.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 1.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 3.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.9 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 1.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 2.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 3.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.7 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 1.0 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.5 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.9 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 4.9 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 4.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.6 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 2.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 5.7 | GO:0008514 | organic anion transmembrane transporter activity(GO:0008514) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 2.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 2.5 | GO:0042277 | peptide binding(GO:0042277) |
| 0.0 | 3.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 3.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.4 | GO:0022884 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 3.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 2.8 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 1.2 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.2 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 2.7 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 1.0 | GO:0003678 | DNA helicase activity(GO:0003678) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 5.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 3.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 1.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 0.6 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 0.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 1.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.9 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.8 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 2.7 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 4.1 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.2 | 3.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 1.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 1.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 1.8 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.2 | 1.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 2.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 2.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 4.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 0.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 4.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 0.9 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 3.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.6 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |