Project
DANIO-CODE
Navigation
Downloads
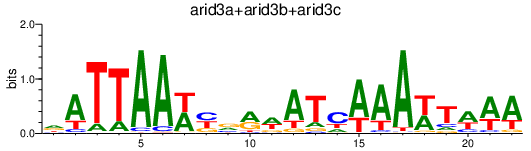
Results for arid3a+arid3b+arid3c
Z-value: 1.35
Transcription factors associated with arid3a+arid3b+arid3c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arid3c
|
ENSDARG00000067729 | AT rich interactive domain 3C (BRIGHT-like) |
|
arid3a
|
ENSDARG00000070843 | AT rich interactive domain 3A (BRIGHT-like) |
|
arid3b
|
ENSDARG00000104034 | AT rich interactive domain 3B (BRIGHT-like) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arid3b | dr10_dc_chr25_-_28630945_28631107 | 0.67 | 4.2e-03 | Click! |
| arid3a | dr10_dc_chr11_+_5745644_5745896 | 0.62 | 9.8e-03 | Click! |
| arid3c | dr10_dc_chr5_+_40722565_40722689 | 0.20 | 4.5e-01 | Click! |
Activity profile of arid3a+arid3b+arid3c motif
Sorted Z-values of arid3a+arid3b+arid3c motif
Network of associatons between targets according to the STRING database.
First level regulatory network of arid3a+arid3b+arid3c
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_3244042 | 7.23 |
ENSDART00000060162
|
hspb1
|
heat shock protein, alpha-crystallin-related, 1 |
| chr5_-_3244261 | 6.41 |
ENSDART00000060162
|
hspb1
|
heat shock protein, alpha-crystallin-related, 1 |
| chr5_+_69383382 | 6.24 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| chr19_-_3226831 | 5.72 |
ENSDART00000145710
ENSDART00000110763 ENSDART00000074620 |
stm
|
starmaker |
| chr19_-_3226933 | 5.03 |
ENSDART00000105174
|
stm
|
starmaker |
| chr17_-_970957 | 3.04 |
ENSDART00000177536
|
dnajc17
|
DnaJ (Hsp40) homolog, subfamily C, member 17 |
| chr19_-_14329389 | 2.43 |
ENSDART00000164594
|
ta
|
T, brachyury homolog a |
| chr1_+_6833994 | 2.07 |
ENSDART00000016139
|
lancl1
|
LanC antibiotic synthetase component C-like 1 (bacterial) |
| chr15_+_37039861 | 1.49 |
ENSDART00000172664
|
kirrel3l
|
kin of IRRE like 3 like |
| chr25_-_10919875 | 1.42 |
ENSDART00000099572
|
mespab
|
mesoderm posterior ab |
| chr19_-_3299610 | 1.41 |
ENSDART00000105168
|
si:ch211-133n4.4
|
si:ch211-133n4.4 |
| chr11_+_10557530 | 1.40 |
ENSDART00000132365
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr15_+_45526576 | 1.30 |
ENSDART00000055978
|
CABZ01067244.1
|
ENSDARG00000020891 |
| chr11_+_42758832 | 1.23 |
|
|
|
| chr22_-_27082972 | 1.20 |
ENSDART00000077411
|
cxcl12b
|
chemokine (C-X-C motif) ligand 12b (stromal cell-derived factor 1) |
| chr11_+_7139675 | 1.15 |
ENSDART00000155864
|
CU929070.1
|
ENSDARG00000097452 |
| chr16_-_45950530 | 1.13 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr10_+_4962403 | 1.11 |
ENSDART00000134679
|
CU074419.2
|
ENSDARG00000093688 |
| chr23_+_32073761 | 1.01 |
|
|
|
| chr16_+_1073570 | 1.01 |
|
|
|
| chr3_-_25144722 | 0.94 |
ENSDART00000055445
|
ddx5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr14_-_11745596 | 0.91 |
ENSDART00000029366
|
zgc:66447
|
zgc:66447 |
| chr3_-_10342940 | 0.90 |
ENSDART00000124419
|
BX539325.2
|
ENSDARG00000101154 |
| chr14_-_32624823 | 0.88 |
ENSDART00000114973
|
cdx4
|
caudal type homeobox 4 |
| chr21_-_11877525 | 0.84 |
ENSDART00000145194
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr7_+_71757218 | 0.80 |
ENSDART00000012918
|
psmd9
|
proteasome 26S subunit, non-ATPase 9 |
| chr3_-_39346621 | 0.79 |
ENSDART00000135192
ENSDART00000013553 ENSDART00000167289 |
zgc:100868
|
zgc:100868 |
| chr5_-_64348528 | 0.76 |
ENSDART00000158856
|
anxa1b
|
annexin A1b |
| chr15_-_47269988 | 0.71 |
ENSDART00000151594
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr2_+_32036450 | 0.70 |
ENSDART00000140776
|
CR391940.1
|
ENSDARG00000091946 |
| chr13_+_27102308 | 0.69 |
ENSDART00000145901
|
rin2
|
Ras and Rab interactor 2 |
| chr12_-_13611954 | 0.67 |
ENSDART00000124638
ENSDART00000124364 |
stat5b
|
signal transducer and activator of transcription 5b |
| chr6_+_54213569 | 0.65 |
ENSDART00000128456
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr25_-_28630138 | 0.65 |
|
|
|
| chr9_+_2472749 | 0.64 |
ENSDART00000147034
|
gpr155a
|
G protein-coupled receptor 155a |
| chr2_-_34010299 | 0.64 |
ENSDART00000140910
|
ptch2
|
patched 2 |
| chr2_+_4233224 | 0.62 |
ENSDART00000163737
|
mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr4_+_17682574 | 0.62 |
ENSDART00000121714
|
gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr20_+_31366832 | 0.60 |
ENSDART00000133353
|
apobb.1
|
apolipoprotein Bb, tandem duplicate 1 |
| chr5_-_24976098 | 0.58 |
ENSDART00000165147
|
ENSDARG00000093253
|
ENSDARG00000093253 |
| chr20_+_53227959 | 0.57 |
|
|
|
| chr18_+_5986137 | 0.56 |
ENSDART00000125725
|
si:ch1073-390k14.1
|
si:ch1073-390k14.1 |
| chr6_+_26957509 | 0.56 |
ENSDART00000153595
|
farp2
|
FERM, RhoGEF and pleckstrin domain protein 2 |
| chr8_-_13562525 | 0.55 |
ENSDART00000139285
|
ENSDARG00000093521
|
ENSDARG00000093521 |
| chr8_-_29921560 | 0.54 |
ENSDART00000125173
|
ercc6l2
|
excision repair cross-complementation group 6-like 2 |
| chr23_+_4841986 | 0.53 |
ENSDART00000135933
|
vgll4a
|
vestigial-like family member 4a |
| chr21_-_43176006 | 0.53 |
ENSDART00000148630
|
hspa4a
|
heat shock protein 4a |
| chr3_+_25888520 | 0.52 |
ENSDART00000135389
|
foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr9_-_56423275 | 0.52 |
|
|
|
| chr11_-_18393802 | 0.52 |
ENSDART00000125453
|
dido1
|
death inducer-obliterator 1 |
| chr16_-_51390400 | 0.52 |
ENSDART00000148894
|
serpinb14
|
serpin peptidase inhibitor, clade B (ovalbumin), member 14 |
| chr25_+_386916 | 0.52 |
|
|
|
| chr20_-_4841337 | 0.50 |
|
|
|
| chr11_-_42096921 | 0.50 |
|
|
|
| chr5_+_68877574 | 0.50 |
ENSDART00000097244
|
gtf2h3
|
general transcription factor IIH, polypeptide 3 |
| chr2_+_53253623 | 0.48 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr13_+_27102377 | 0.48 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr5_+_71099904 | 0.48 |
ENSDART00000170215
|
nup214
|
nucleoporin 214 |
| chr15_+_42329269 | 0.47 |
ENSDART00000099234
ENSDART00000152731 |
scaf4b
|
SR-related CTD-associated factor 4b |
| chr8_+_456029 | 0.47 |
ENSDART00000051776
|
srp19
|
signal recognition particle 19 |
| chr10_+_44738199 | 0.46 |
|
|
|
| chr22_+_38096040 | 0.46 |
ENSDART00000097533
|
wwtr1
|
WW domain containing transcription regulator 1 |
| chr24_+_26287471 | 0.46 |
ENSDART00000105784
ENSDART00000122554 |
cldn11b
|
claudin 11b |
| chr7_-_8465988 | 0.46 |
ENSDART00000172928
|
tex261
|
testis expressed 261 |
| chr22_-_10612143 | 0.42 |
ENSDART00000064772
|
cyb561d2
|
cytochrome b561 family, member D2 |
| chr23_-_24307705 | 0.42 |
ENSDART00000124539
|
ddost
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr4_+_24006115 | 0.42 |
|
|
|
| chr9_+_2523927 | 0.41 |
ENSDART00000166326
|
si:ch73-167c12.2
|
si:ch73-167c12.2 |
| chr20_+_21002097 | 0.41 |
ENSDART00000035827
|
brf1b
|
BRF1, RNA polymerase III transcription initiation factor b |
| chr8_+_48976657 | 0.41 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr25_-_16658906 | 0.41 |
ENSDART00000124729
ENSDART00000110859 |
ribc2
|
RIB43A domain with coiled-coils 2 |
| chr9_-_34491458 | 0.41 |
ENSDART00000049805
|
ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr4_-_5766814 | 0.40 |
ENSDART00000021753
|
ccnc
|
cyclin C |
| chr3_+_42293213 | 0.40 |
ENSDART00000163595
|
ints1
|
integrator complex subunit 1 |
| chr16_-_21334269 | 0.40 |
ENSDART00000145837
|
si:dkey-271j15.3
|
si:dkey-271j15.3 |
| chr9_+_3458086 | 0.39 |
ENSDART00000160977
ENSDART00000114168 |
CU469503.1
itga6a
|
ENSDARG00000099348 integrin, alpha 6a |
| chr1_-_11474269 | 0.38 |
ENSDART00000054801
|
b4galt1
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
| chr24_+_36451347 | 0.38 |
ENSDART00000142264
|
grnb
|
granulin b |
| chr21_-_39900518 | 0.37 |
ENSDART00000175055
|
CABZ01065291.1
|
ENSDARG00000107374 |
| chr19_-_41243536 | 0.37 |
|
|
|
| chr14_-_943860 | 0.36 |
ENSDART00000010773
|
acsl1b
|
acyl-CoA synthetase long-chain family member 1b |
| chr18_-_21921898 | 0.35 |
ENSDART00000132381
|
edc4
|
enhancer of mRNA decapping 4 |
| chr1_-_21987718 | 0.35 |
ENSDART00000128918
|
fgfbp1b
|
fibroblast growth factor binding protein 1b |
| chr22_+_39067994 | 0.34 |
|
|
|
| chr23_+_4842025 | 0.34 |
ENSDART00000135933
|
vgll4a
|
vestigial-like family member 4a |
| chr8_-_46313975 | 0.34 |
ENSDART00000075189
|
mtor
|
mechanistic target of rapamycin (serine/threonine kinase) |
| chr6_+_49552893 | 0.34 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr16_-_10425382 | 0.33 |
ENSDART00000104025
ENSDART00000178832 |
flot1b
|
flotillin 1b |
| chr3_-_36126124 | 0.33 |
|
|
|
| chr8_+_6989445 | 0.32 |
ENSDART00000134440
|
gpd1a
|
glycerol-3-phosphate dehydrogenase 1a |
| chr1_-_11474192 | 0.32 |
ENSDART00000054801
|
b4galt1
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
| chr10_+_42530040 | 0.32 |
ENSDART00000025691
|
dbnla
|
drebrin-like a |
| chr19_+_26386711 | 0.32 |
ENSDART00000089836
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr8_+_47108500 | 0.32 |
ENSDART00000142979
|
arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr9_-_23340795 | 0.32 |
ENSDART00000133017
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr6_-_42371368 | 0.32 |
ENSDART00000039868
|
usp4
|
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr5_+_29114616 | 0.31 |
ENSDART00000148314
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr14_+_47028640 | 0.31 |
|
|
|
| chr10_-_43924675 | 0.30 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr15_+_27008152 | 0.30 |
ENSDART00000077172
|
bcas3
|
breast carcinoma amplified sequence 3 |
| chr14_-_21717949 | 0.30 |
ENSDART00000137795
|
ssrp1a
|
structure specific recognition protein 1a |
| chr1_-_11474337 | 0.29 |
ENSDART00000149913
|
b4galt1
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
| chr5_+_50252203 | 0.29 |
ENSDART00000170756
ENSDART00000083317 |
arsk
|
arylsulfatase family, member K |
| chr25_-_20933295 | 0.29 |
ENSDART00000138985
ENSDART00000046298 |
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr14_+_35880163 | 0.29 |
ENSDART00000148319
|
pitx2
|
paired-like homeodomain 2 |
| chr12_-_944349 | 0.28 |
ENSDART00000128188
|
daglb
|
diacylglycerol lipase, beta |
| chr12_-_33558313 | 0.28 |
ENSDART00000111259
|
tmem94
|
transmembrane protein 94 |
| chr22_+_2849533 | 0.28 |
ENSDART00000106620
|
zgc:174224
|
zgc:174224 |
| chr21_-_37404394 | 0.27 |
ENSDART00000129439
|
BX649250.1
|
ENSDARG00000091187 |
| chr9_+_2472639 | 0.27 |
ENSDART00000147034
|
gpr155a
|
G protein-coupled receptor 155a |
| chr18_+_13267658 | 0.26 |
ENSDART00000091560
|
tldc1
|
TBC/LysM-associated domain containing 1 |
| chr8_+_23763721 | 0.26 |
ENSDART00000062968
|
si:ch211-163l21.8
|
si:ch211-163l21.8 |
| chr19_+_43537350 | 0.26 |
|
|
|
| chr21_+_6291936 | 0.25 |
ENSDART00000136539
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr2_+_21067708 | 0.25 |
ENSDART00000148400
ENSDART00000021168 |
rxrga
|
retinoid x receptor, gamma a |
| chr17_+_989918 | 0.25 |
ENSDART00000169903
|
CABZ01060373.1
|
ENSDARG00000103671 |
| chr23_-_25028527 | 0.24 |
ENSDART00000144903
|
zbtb48
|
zinc finger and BTB domain containing 48 |
| chr4_-_13932592 | 0.24 |
ENSDART00000067174
|
zcrb1
|
zinc finger CCHC-type and RNA binding motif 1 |
| chr6_+_55835894 | 0.24 |
ENSDART00000108786
|
si:ch211-81n22.1
|
si:ch211-81n22.1 |
| chr19_-_3186346 | 0.24 |
ENSDART00000146928
|
si:ch211-80h18.1
|
si:ch211-80h18.1 |
| chr20_-_4841465 | 0.23 |
|
|
|
| chr9_-_34459378 | 0.23 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr16_+_20361962 | 0.23 |
ENSDART00000004031
|
hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr10_+_5413893 | 0.23 |
ENSDART00000138821
|
nfil3
|
nuclear factor, interleukin 3 regulated |
| chr11_+_16439987 | 0.22 |
ENSDART00000104072
|
kbtbd8
|
kelch repeat and BTB (POZ) domain containing 8 |
| chr5_+_66712197 | 0.22 |
ENSDART00000014822
|
ebf2
|
early B-cell factor 2 |
| chr1_+_24960066 | 0.22 |
ENSDART00000054235
|
plrg1
|
pleiotropic regulator 1 |
| chr24_-_25099449 | 0.22 |
ENSDART00000153798
|
hhla2b.2
|
HERV-H LTR-associating 2b, tandem duplicate 2 |
| chr13_+_5850284 | 0.21 |
ENSDART00000121598
|
phf10
|
PHD finger protein 10 |
| chr12_-_33558248 | 0.21 |
ENSDART00000153457
|
tmem94
|
transmembrane protein 94 |
| chr1_-_2301566 | 0.21 |
ENSDART00000103785
|
ggact.3
|
gamma-glutamylamine cyclotransferase, tandem duplicate 3 |
| chr16_-_4780901 | 0.21 |
ENSDART00000029485
|
arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr16_+_17705704 | 0.21 |
|
|
|
| chr2_-_9691551 | 0.21 |
ENSDART00000146715
|
st6galnac3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr6_+_9441851 | 0.21 |
ENSDART00000064995
|
sumo1
|
small ubiquitin-like modifier 1 |
| chr19_-_2314582 | 0.21 |
ENSDART00000166669
|
btd
|
biotinidase |
| chr20_-_28531019 | 0.20 |
ENSDART00000172133
|
CABZ01057122.1
|
ENSDARG00000105192 |
| chr2_-_14833347 | 0.20 |
|
|
|
| chr23_-_29931398 | 0.20 |
ENSDART00000109506
|
tmem201
|
transmembrane protein 201 |
| chr23_+_22649652 | 0.20 |
ENSDART00000146709
|
CR388207.1
|
ENSDARG00000095377 |
| chr7_+_36195907 | 0.20 |
ENSDART00000138893
|
aktip
|
akt interacting protein |
| chr20_-_54206786 | 0.20 |
ENSDART00000004756
|
hsp90aa1.1
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 1 |
| chr5_-_9162684 | 0.20 |
|
|
|
| chr5_-_64149931 | 0.19 |
ENSDART00000144816
|
lix1
|
limb and CNS expressed 1 |
| chr19_-_46427187 | 0.19 |
ENSDART00000169957
|
krit1
|
KRIT1, ankyrin repeat containing |
| chr20_-_873476 | 0.19 |
ENSDART00000142361
ENSDART00000104725 |
snx14
|
sorting nexin 14 |
| chr15_-_3547167 | 0.19 |
ENSDART00000162677
|
cog6
|
component of oligomeric golgi complex 6 |
| chr5_+_45677439 | 0.19 |
ENSDART00000045598
|
zgc:110626
|
zgc:110626 |
| chr18_+_20493237 | 0.19 |
ENSDART00000128139
|
kbtbd4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr10_+_45184723 | 0.19 |
ENSDART00000160536
|
gnsb
|
glucosamine (N-acetyl)-6-sulfatase (Sanfilippo disease IIID), b |
| chr23_+_4842211 | 0.18 |
ENSDART00000171971
|
vgll4a
|
vestigial-like family member 4a |
| chr9_+_7832253 | 0.18 |
ENSDART00000109288
|
myo16
|
myosin XVI |
| chr12_-_48288906 | 0.18 |
ENSDART00000171058
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr6_+_58704352 | 0.18 |
ENSDART00000172466
|
igsf8
|
immunoglobulin superfamily, member 8 |
| chr3_-_55278137 | 0.18 |
ENSDART00000123544
|
tex2
|
testis expressed 2 |
| chr11_-_864648 | 0.18 |
ENSDART00000162152
|
atg7
|
ATG7 autophagy related 7 homolog (S. cerevisiae) |
| chr6_-_23830912 | 0.17 |
|
|
|
| chr11_-_31012985 | 0.17 |
ENSDART00000162605
|
man2b1
|
mannosidase, alpha, class 2B, member 1 |
| chr3_+_25247071 | 0.17 |
|
|
|
| chr17_-_36988937 | 0.17 |
ENSDART00000145236
|
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr10_-_20748720 | 0.17 |
ENSDART00000064581
|
kcnip3b
|
Kv channel interacting protein 3b, calsenilin |
| chr8_-_455930 | 0.16 |
ENSDART00000051777
|
zgc:101664
|
zgc:101664 |
| chr2_-_14833381 | 0.16 |
|
|
|
| chr21_-_33196706 | 0.16 |
ENSDART00000003983
|
rbm22
|
RNA binding motif protein 22 |
| chr5_+_23409703 | 0.16 |
ENSDART00000015401
|
ercc6l
|
excision repair cross-complementation group 6-like |
| chr21_+_25199691 | 0.16 |
ENSDART00000168140
ENSDART00000112783 |
tmem45b
|
transmembrane protein 45B |
| chr14_-_6738260 | 0.16 |
ENSDART00000171792
|
rufy1
|
RUN and FYVE domain containing 1 |
| chr1_-_50576110 | 0.15 |
ENSDART00000146612
|
esf1
|
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae) |
| chr24_+_9605832 | 0.15 |
ENSDART00000131891
|
tmem108
|
transmembrane protein 108 |
| chr11_-_865163 | 0.15 |
ENSDART00000172904
|
atg7
|
ATG7 autophagy related 7 homolog (S. cerevisiae) |
| chr12_-_29762589 | 0.15 |
|
|
|
| chr6_+_56092642 | 0.15 |
ENSDART00000059438
|
rtfdc1
|
replication termination factor 2 domain containing 1 |
| chr8_+_48976926 | 0.15 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr12_+_2835894 | 0.15 |
ENSDART00000165225
|
prkar1ab
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) b |
| chr23_-_29627060 | 0.15 |
ENSDART00000166554
|
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr1_+_56554227 | 0.15 |
ENSDART00000149688
|
mycbpap
|
mycbp associated protein |
| chr18_-_3244618 | 0.15 |
ENSDART00000161520
|
gdpd4a
|
glycerophosphodiester phosphodiesterase domain containing 4a |
| chr2_+_22875548 | 0.14 |
ENSDART00000157884
|
hfm1
|
HFM1, ATP-dependent DNA helicase homolog (S. cerevisiae) |
| chr7_-_59851104 | 0.14 |
ENSDART00000128363
|
cct7
|
chaperonin containing TCP1, subunit 7 (eta) |
| chr23_+_25028596 | 0.14 |
ENSDART00000142124
|
nol9
|
nucleolar protein 9 |
| chr11_-_25223723 | 0.14 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr6_+_4379989 | 0.14 |
ENSDART00000025031
|
pou4f1
|
POU class 4 homeobox 1 |
| chr1_+_1564283 | 0.14 |
ENSDART00000166968
|
atp1a1a.2
|
ATPase, Na+/K+ transporting, alpha 1a polypeptide, tandem duplicate 2 |
| chr10_-_40904685 | 0.14 |
|
|
|
| chr18_+_35198982 | 0.14 |
ENSDART00000127379
|
cfap45
|
cilia and flagella associated protein 45 |
| chr5_+_65754237 | 0.14 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr15_-_1768269 | 0.14 |
ENSDART00000154668
|
rabgef1l
|
RAB guanine nucleotide exchange factor (GEF) 1, like |
| chr17_+_22291546 | 0.14 |
ENSDART00000151929
ENSDART00000089919 ENSDART00000000804 |
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr1_+_31608918 | 0.13 |
ENSDART00000137156
|
pudp
|
pseudouridine 5'-phosphatase |
| chr1_+_232001 | 0.13 |
ENSDART00000003317
|
tfdp1b
|
transcription factor Dp-1, b |
| chr15_-_19186046 | 0.13 |
ENSDART00000108818
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr4_-_33510045 | 0.13 |
ENSDART00000150625
|
si:dkeyp-4f2.3
|
si:dkeyp-4f2.3 |
| chr24_-_37596370 | 0.13 |
ENSDART00000162538
|
cluap1
|
clusterin associated protein 1 |
| chr2_-_50537722 | 0.13 |
ENSDART00000127623
|
cul1a
|
cullin 1a |
| chr14_-_24463906 | 0.13 |
ENSDART00000126199
|
slit3
|
slit homolog 3 (Drosophila) |
| chr18_+_35198798 | 0.13 |
ENSDART00000098292
|
cfap45
|
cilia and flagella associated protein 45 |
| chr10_+_44738165 | 0.12 |
|
|
|
| chr2_+_1615812 | 0.12 |
ENSDART00000113068
|
mcoln3a
|
mucolipin 3a |
| chr23_-_19898781 | 0.12 |
ENSDART00000144027
ENSDART00000139425 |
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr8_+_22372084 | 0.12 |
ENSDART00000164143
|
zgc:153759
|
zgc:153759 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 13.6 | GO:0046685 | regulation of myofibril size(GO:0014881) response to arsenic-containing substance(GO:0046685) |
| 1.8 | 10.8 | GO:0045299 | otolith mineralization(GO:0045299) |
| 1.0 | 6.2 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) |
| 0.3 | 0.9 | GO:0002824 | positive regulation of adaptive immune response(GO:0002821) positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains(GO:0002824) |
| 0.3 | 1.2 | GO:0097534 | trigeminal sensory nucleus development(GO:0021730) lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.2 | 0.7 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.2 | 1.0 | GO:0002090 | regulation of receptor internalization(GO:0002090) |
| 0.1 | 0.9 | GO:0003311 | pancreatic D cell differentiation(GO:0003311) foregut morphogenesis(GO:0007440) |
| 0.1 | 0.6 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.1 | 0.3 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.1 | 1.4 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 0.7 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.5 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 1.5 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.3 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 0.3 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 0.2 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 0.4 | GO:0000423 | macromitophagy(GO:0000423) C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.2 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 0.3 | GO:0061303 | iris morphogenesis(GO:0061072) cornea development in camera-type eye(GO:0061303) |
| 0.1 | 0.4 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.6 | GO:0002063 | chondrocyte development(GO:0002063) protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.3 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.2 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 0.3 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.1 | 0.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.3 | GO:0071926 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.1 | 0.6 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.2 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.0 | 1.1 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.6 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 1.0 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.6 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.2 | GO:0048769 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) sarcomerogenesis(GO:0048769) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.4 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.4 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.5 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.5 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.4 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0046950 | cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.0 | 0.1 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.2 | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043516) |
| 0.0 | 0.1 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 0.5 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.0 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.2 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.1 | GO:0032655 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.0 | 0.0 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.2 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.2 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.0 | 0.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.8 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.6 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.2 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.5 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 0.8 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.5 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.4 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 13.1 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 13.3 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.7 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.2 | 1.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 1.0 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.2 | 0.6 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.1 | 1.2 | GO:0008009 | chemokine activity(GO:0008009) chemokine receptor binding(GO:0042379) |
| 0.1 | 0.4 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 0.9 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.1 | 0.3 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.4 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.1 | 0.5 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.1 | 0.6 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.1 | 0.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.3 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 0.4 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.4 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.4 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.3 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.8 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.5 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.3 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.6 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.5 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 13.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 0.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 0.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.3 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 14.5 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 0.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.0 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.5 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |